4NIW
 
 | |
4NFF
 
 | | Human kallikrein-related peptidase 2 in complex with PPACK | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Kallikrein-2 | | Authors: | Skala, W, Brandstetter, H, Magdolen, V, Goettig, P. | | Deposit date: | 2013-10-31 | | Release date: | 2014-10-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity
J.Biol.Chem., 289, 2014
|
|
4NIX
 
 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) orthorhombic form, zinc-bound | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NIV
 
 | |
8OXV
 
 | |
8OWU
 
 | |
8OWS
 
 | |
7API
 
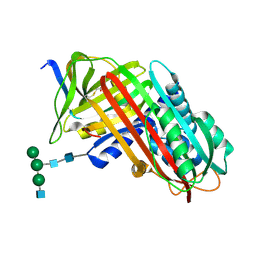 | | THE S VARIANT OF HUMAN ALPHA1-ANTITRYPSIN, STRUCTURE AND IMPLICATIONS FOR FUNCTION AND METABOLISM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTITRYPSIN, ... | | Authors: | Loebermann, H, Tokuoka, R, Deisenhofer, J, Huber, R. | | Deposit date: | 1988-09-08 | | Release date: | 1990-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism.
Protein Eng., 2, 1989
|
|
2BHG
 
 | |
2FYR
 
 | |
8GZ9
 
 | | Cryo-EM structure of Abeta2 fibril polymorph2 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
8GZ8
 
 | | Cryo-EM structure of Abeta2 fibril polymorph1 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
2MZE
 
 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) | | Descriptor: | CALCIUM ION, Matrilysin, ZINC ION | | Authors: | Prior, S.H, Fulcher, Y.G, Van Doren, S.R. | | Deposit date: | 2015-02-11 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
6S58
 
 | | AvaII restriction endonuclease in the absence of nucleic acids | | Descriptor: | CALCIUM ION, Type II site-specific deoxyribonuclease, UNKNOWN ATOM OR ION | | Authors: | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | Deposit date: | 2019-06-30 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
6RGV
 
 | |
6S48
 
 | | AvaII RESTRICTION ENDONUCLEASE IN COMPLEX WITH PARTIALLY CLEAVED dsDNA | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, DNA (5'-D(*GP*AP*TP*G)-3'), ... | | Authors: | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | Deposit date: | 2019-06-26 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
2MZI
 
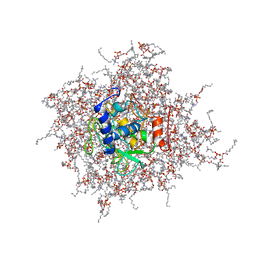 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) in Complex with Anionic Membrane | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLEST-5-EN-3-YL HYDROGEN SULFATE, ... | | Authors: | Prior, S.H, Van Doren, S.R. | | Deposit date: | 2015-02-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
7N2G
 
 | | MicroED structure of human CPEB3 segment(154-161) kinked polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.201 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2F
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2E
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
1JXC
 
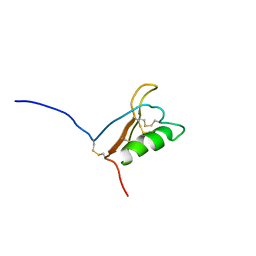 | | Minimized NMR structure of ATT, an Arabidopsis trypsin/chymotrypsin inhibitor | | Descriptor: | Putative trypsin inhibitor ATTI-2 | | Authors: | Zhao, Q, Chae, Y.K, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2001-09-06 | | Release date: | 2003-01-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of ATTp, an Arabidopsis thaliana Trypsin Inhibitor
Biochemistry, 41, 2002
|
|
5O4X
 
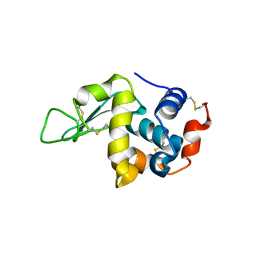 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | Descriptor: | Lysozyme C | | Authors: | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | Cite: | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2MZH
 
 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) in Complex with Zwitterionic Membrane | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Matrilysin, ... | | Authors: | Prior, S.H, Van Doren, S.R. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
1KIO
 
 | | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGCI[L30R, K31M] | | Descriptor: | SERINE PROTEASE INHIBITOR I | | Authors: | Gaspari, Z, Patthy, A, Graf, L, Perczel, A. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
Eur.J.Biochem., 269, 2002
|
|
7NS7
 
 | |
