1QO2
 
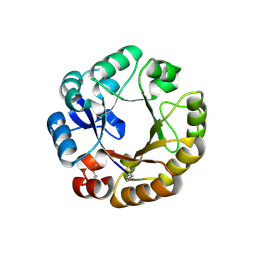 | | Crystal structure of N-((5'-phosphoribosyl)-formimino)-5-aminoimidazol-4-carboxamid ribonucleotid isomerase (EC 3.1.3.15, HisA) | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase | | Authors: | Wilmanns, M, Lang, D, Thoma, R, Sterner, R. | | Deposit date: | 1999-11-01 | | Release date: | 2000-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Evidence for Evolution of the Beta/Alpha-Barrel Scaffold by Repeated Gene Duplication and Fusion
Science, 289, 2000
|
|
1QO3
 
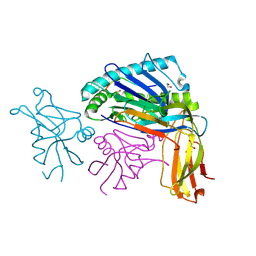 | |
1QO4
 
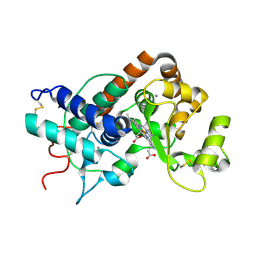 | | ARABIDOPSIS THALIANA PEROXIDASE A2 AT ROOM TEMPERATURE | | Descriptor: | CALCIUM ION, PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Henriksen, A, Gajhede, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Activity and Structure of Highly Similar Peroxidases. Spectroscopic, Crystallographic, and Enzymatic Analyses of Lignifying Arabidopsis Thaliana Peroxidase A2 and Horseradish Peroxidase A2
Biochemistry, 40, 2001
|
|
1QO5
 
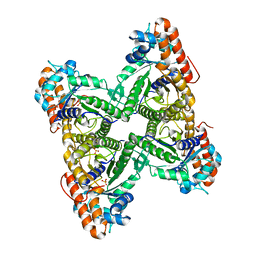 | |
1QO6
 
 | |
1QO7
 
 | | Structure of Aspergillus niger epoxide hydrolase | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Zou, J.-Y, Hallberg, B.M, Bergfors, T, Oesch, F, Arand, M, Mowbray, S.L, Jones, T.A. | | Deposit date: | 1999-11-04 | | Release date: | 2000-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Aspergillus Niger Epoxide Hydrolase at 1.8A Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases
Structure, 8, 2000
|
|
1QO8
 
 | | The structure of the open conformation of a flavocytochrome c3 fumarate reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C3 FUMARATE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bamford, V, Dobbin, P.S, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 1999-11-04 | | Release date: | 2000-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Open Conformation of a Flavocytochrome C3 Fumarate Reductase.
Nat.Struct.Biol., 6, 1999
|
|
1QOA
 
 | | FERREDOXIN MUTATION C49S | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron-sulfur cluster cysteine-to-serine mutants of Anabaena -2Fe-2S- ferredoxin exhibit unexpected redox properties and are competent in electron transfer to ferredoxin:NADP+ reductase.
Biochemistry, 36, 1997
|
|
1QOB
 
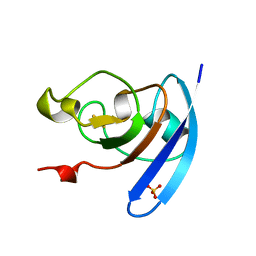 | | FERREDOXIN MUTATION D62K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOF
 
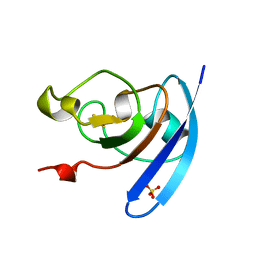 | | FERREDOXIN MUTATION Q70K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOG
 
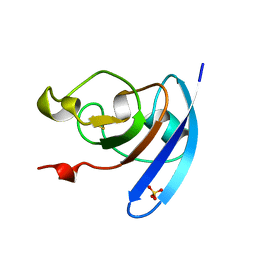 | | FERREDOXIN MUTATION S47A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOH
 
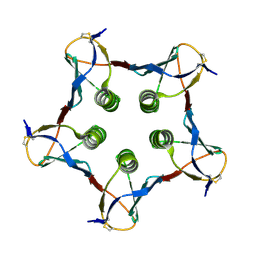 | | A MUTANT SHIGA-LIKE TOXIN IIE | | Descriptor: | SHIGA-LIKE TOXIN IIE B SUBUNIT | | Authors: | Pannu, N.S, Boodhoo, A, Armstrong, G.D, Clark, C.G, Brunton, J.L, Read, R.J. | | Deposit date: | 1999-11-08 | | Release date: | 2000-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Mutant Shiga-Like Toxin Iie Bound to its Receptor Gb(3): Structure of a Group II Shiga-Like Toxin with Altered Binding Specificity
Structure, 8, 2000
|
|
1QOI
 
 | | U4/U6 snRNP-specific cyclophilin SnuCyp-20 | | Descriptor: | SNUCYP-20 | | Authors: | Reidt, U, Reuter, K, Achsel, T, Ingelfinger, D, Luehrmann, R, Ficner, R. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human U4/U6 Small Nuclear Ribonucleoproteinparticle-Specificsnucyp-20, a Nuclear Cyclophilin
J.Biol.Chem., 275, 2000
|
|
1QOJ
 
 | | Crystal Structure of E.coli UvrB C-terminal domain, and a model for UvrB-UvrC interaction. | | Descriptor: | UVRB | | Authors: | Sohi, M, Alexandrovich, A, Moolenaar, G, Visse, R, Goosen, N, Vernede, X, Fontecilla-Camps, J, Champness, J, Sanderson, M.R. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-10 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of E.Coli Uvrb C-Terminal Domain, and a Model for Uvrb-Uvrc Interaction
FEBS Lett., 465, 2000
|
|
1QOK
 
 | | MFE-23 AN ANTI-CARCINOEMBRYONIC ANTIGEN SINGLE-CHAIN FV ANTIBODY | | Descriptor: | MFE-23 RECOMBINANT ANTIBODY FRAGMENT | | Authors: | Boehm, M.K, Corper, A.L, Wan, T, Sohi, M.K, Sutton, B.J, Thornton, J.D, Keep, P.A, Chester, K.A, Begent, R.H.J, Perkins, S.J. | | Deposit date: | 1999-11-11 | | Release date: | 2000-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Anti-Carcinoembryonic Antigen Single-Chain Fv Antibody Mfe-23 and a Model for Antigen Binding Based on Intermolecular Contacts
Biochem.J., 346, 2000
|
|
1QOL
 
 | | STRUCTURE OF THE FMDV LEADER PROTEASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROTEASE (NONSTRUCTURAL PROTEIN P20A) | | Authors: | Guarne, A, Tormo, J, Kirchweger, R, Pfistermueller, D, Skern, T, Fita, I. | | Deposit date: | 1999-11-13 | | Release date: | 2000-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Foot-and-Mouth Disease Virus Leader Protease: A Papain-Like Fold Adapted for Self-Processing and Eif4G Recognition.
Embo J., 17, 1998
|
|
1QOM
 
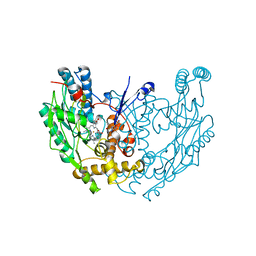 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH SWAPPED N-TERMINAL HOOK | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Crane, B.R, Rosenfeld, R.A, Arvai, A.S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-15 | | Release date: | 1999-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-Terminal Domain Swapping and Metal Ion Binding in Nitric Oxide Synthase Dimerization
Embo J., 18, 1999
|
|
1QOO
 
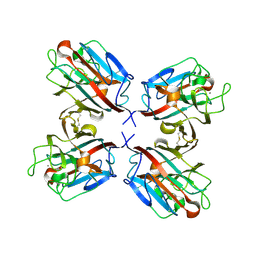 | | lectin UEA-II complexed with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1QOP
 
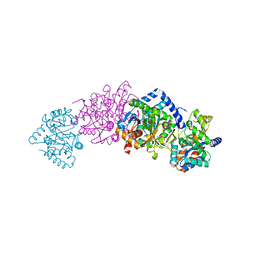 | |
1QOQ
 
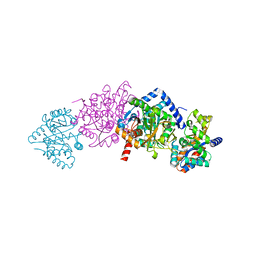 | |
1QOR
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI QUINONE OXIDOREDUCTASE COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, QUINONE OXIDOREDUCTASE, SULFATE ION | | Authors: | Thorn, J.M, Barton, J.D, Dixon, N.E, Ollis, D.L, Edwards, K.J. | | Deposit date: | 1995-02-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH.
J.Mol.Biol., 249, 1995
|
|
1QOS
 
 | | lectin UEA-II complexed with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-16 | | Release date: | 2000-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1QOT
 
 | | lectin UEA-II complexed with fucosyllactose and fucosylgalactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-16 | | Release date: | 1999-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1QOU
 
 | |
1QOV
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M260 REPLACED WITH TRP (CHAIN M, A260W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | McAuley, K.E, Fyfe, P.K, Ridge, J.P, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 1999-11-17 | | Release date: | 1999-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Details of an Interaction between Cardiolipin and an Integral Membrane Protein
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
