1QL6
 
 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
1QL7
 
 | | FACTOR XA SPECIFIC INHIBITOR IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 1999-08-24 | | Release date: | 2000-08-25 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ph-Dependent Binding Modes Observed in Trypsin Crystals: Lessons for the Structure-Based Drug Design
Chembiochem, 3, 2002
|
|
1QL8
 
 | |
1QL9
 
 | |
1QLB
 
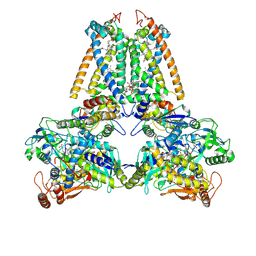 | | respiratory complex II-like fumarate reductase from Wolinella succinogenes | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Lancaster, C.R.D, Kroeger, A, Auer, M, Michel, H. | | Deposit date: | 1999-08-25 | | Release date: | 1999-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of Fumarate Reductase from Wolinella Succinogenes at 2.2 Angstroms Resolution
Nature, 402, 1999
|
|
1QLC
 
 | |
1QLD
 
 | |
1QLE
 
 | |
1QLF
 
 | | MHC CLASS I H-2DB COMPLEXED WITH GLYCOPEPTIDE K3G | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, HUMAN BETA-2-MICROGLOBULIN, ... | | Authors: | Tormo, J, Jones, E.Y. | | Deposit date: | 1999-08-30 | | Release date: | 1999-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Two H-2Db/Glycopeptide Complexes Suggest a Molecular Basis for Ctl Cross-Reactivity
Immunity, 10, 1999
|
|
1QLG
 
 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | Descriptor: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-31 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
1QLH
 
 | |
1QLI
 
 | | QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYSTEINE AND GLYCINE-RICH PROTEIN, ZINC ION | | Authors: | Konrat, R, Weiskirchen, R, Krautler, B, Bister, K. | | Deposit date: | 1997-02-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carboxyl-terminal LIM domain from quail cysteine-rich protein CRP2.
J.Biol.Chem., 272, 1997
|
|
1QLJ
 
 | |
1QLK
 
 | | SOLUTION STRUCTURE OF CA(2+)-LOADED RAT S100B (BETABETA) NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Drohat, A.C, Baldisseri, D.M, Rustandi, R.R, Weber, D.J. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy,.
Biochemistry, 37, 1998
|
|
1QLL
 
 | | Piratoxin-II (Prtx-II) - a K49 PLA2 from Bothrops pirajai | | Descriptor: | N-TRIDECANOIC ACID, PHOSPHOLIPASE A2 | | Authors: | Lee, W.-H, Polikarpov, I. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Low Catalytic Activity in Lys49 Phospholipases A2-A Hypothesis: The Crystal Structure of Piratoxin II Complexed to Fatty Acid
Biochemistry, 40, 2001
|
|
1QLM
 
 | |
1QLN
 
 | | STRUCTURE OF A TRANSCRIBING T7 RNA POLYMERASE INITIATION COMPLEX | | Descriptor: | BACTERIOPHAGE T7 RNA POLYMERASE, DNA (5- D (P*CP*TP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP* AP*TP*TP*A)-3), DNA (5-D(P*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*TP*A)-3), ... | | Authors: | Cheetham, G.M.T, Steitz, T.A. | | Deposit date: | 1999-09-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Transcribing T7 RNA Polymerase Initiation Complex
Science, 286, 1999
|
|
1QLO
 
 | | Structure of the active domain of the herpes simplex virus protein ICP47 in water/sodium dodecyl sulfate solution determined by nuclear magnetic resonance spectroscopy | | Descriptor: | HERPES SIMPLEX VIRUS PROTEIN ICP47 | | Authors: | Pfaender, R, Neumann, L, Zweckstetter, M, Seger, C, Holak, T.A, Tampe, R. | | Deposit date: | 1999-09-09 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Active Domain of the Herpes Simplex Virus Protein Icp47 in Water/Sodium Dodecyl Sulfate Solution Determined by Nuclear Magnetic Resonance Spectroscopy.
Biochemistry, 38, 1999
|
|
1QLP
 
 | | 2.0 ANGSTROM STRUCTURE OF INTACT ALPHA-1-ANTITRYPSIN: A CANONICAL TEMPLATE FOR ACTIVE SERPINS | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Elliott, P.R, Pei, X.Y, Dafforn, T, Read, R.J, Carrell, R.W, Lomas, D.A. | | Deposit date: | 1999-09-10 | | Release date: | 1999-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Topography of a 2.0 A structure of alpha1-antitrypsin reveals targets for rational drug design to prevent conformational disease.
Protein Sci., 9, 2000
|
|
1QLQ
 
 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
1QLR
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A HUMAN MONOCLONAL IgM COLD AGGLUTININ | | Descriptor: | IGM FAB REGION IV-J(H4)-C (KAU COLD AGGLUTININ), IGM KAPPA CHAIN V-III (KAU COLD AGGLUTININ), alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carvalho, J.G, Cauerhff, A, Goldbaum, F, Leoni, J, Polikarpov, I. | | Deposit date: | 1999-09-11 | | Release date: | 2000-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Three-dimensional structure of the Fab from a human IgM cold agglutinin.
J Immunol., 165, 2000
|
|
1QLS
 
 | | S100C (S100A11),OR CALGIZZARIN, IN COMPLEX WITH ANNEXIN I N-TERMINUS | | Descriptor: | ANNEXIN I, CALCIUM ION, S100C PROTEIN | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1999-09-15 | | Release date: | 2000-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Ca2+ Dependent Association between S100C (S100A11) and its Target, the N-Terminal Part of Annexin I
Structure, 8, 2000
|
|
1QLT
 
 | |
1QLU
 
 | |
1QLV
 
 | |
