8WQB
 
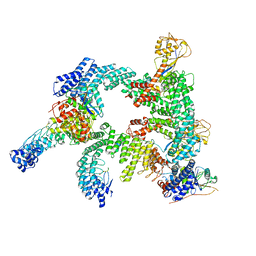 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQA
 
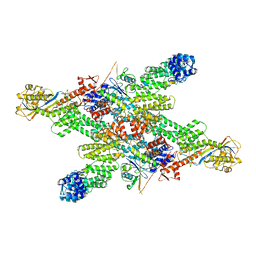 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQH
 
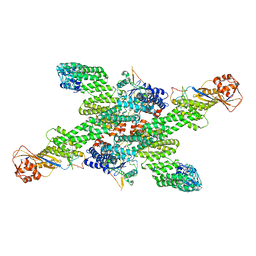 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
4C9Z
 
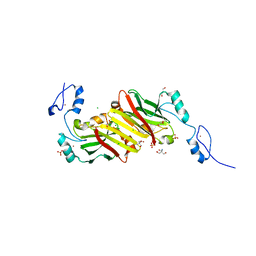 | | Crystal structure of Siah1 at 1.95 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE SIAH1, ... | | Authors: | Rimsa, V, Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-10-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two High-Resolution Structures of the Human E3 Ubiquitin Ligase Siah1.
Acta Crystallogr.,Sect.F, 96, 2013
|
|
4PHY
 
 | | Functional conservation despite structural divergence in ligand-responsive RNA switches | | Descriptor: | ACETATE ION, MAGNESIUM ION, RNA (26-MER), ... | | Authors: | Boerneke, M.A, Dibrov, S.M, Hermann, T.H. | | Deposit date: | 2014-05-07 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional conservation despite structural divergence in ligand-responsive RNA switches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7NRV
 
 | | Paired helical filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRX
 
 | | Straight filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NPC
 
 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM156 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, GLYCEROL, Nuclear receptor ROR-gamma | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
7NP5
 
 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM216 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methoxy]-2-fluoranyl-benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Oerlemans, G.J.M, Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
6EW8
 
 | |
6UEC
 
 | | Pseudomonas aeruginosa LpxD Complex Structure with Ligand | | Descriptor: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
7GY4
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 1.14 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
2ZXM
 
 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Ikura, T, Ito, N. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
4P97
 
 | |
6VOD
 
 | | HIV-1 wild type protease with GRL-052-16A, a tricyclic cyclohexane fused tetrahydrofuranofuran (CHf-THF) derivative as the P2 ligand | | Descriptor: | (1R,3aS,5R,6S,7aR)-octahydro-1,6-epoxy-2-benzofuran-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure-Based Design of Highly Potent HIV-1 Protease Inhibitors Containing New Tricyclic Ring P2-Ligands: Design, Synthesis, Biological, and X-ray Structural Studies.
J.Med.Chem., 63, 2020
|
|
2OH8
 
 | | Myoglobin cavity mutant I28W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
2OHA
 
 | | Myoglobin cavity mutant F138W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
3PAR
 
 | | Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) in the absence of ligand | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A, SULFATE ION | | Authors: | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
2ZXN
 
 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2S,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Ikura, T, Ito, N. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
2Z01
 
 | | Crystal structure of phosphoribosylaminoimidazole synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 2015
|
|
1K4W
 
 | | X-ray structure of the orphan nuclear receptor ROR beta ligand-binding domain in the active conformation | | Descriptor: | Nuclear receptor ROR-beta, STEARIC ACID, steroid receptor coactivator-1 | | Authors: | Stehlin, C, Wurtz, J.M, Steinmetz, A, Greiner, E, Schuele, R, Moras, D, Renaud, J.P. | | Deposit date: | 2001-10-09 | | Release date: | 2002-04-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation.
EMBO J., 20, 2001
|
|
7GV5
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 17.50 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GVC
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 3 at 8.70 MGy X-ray dose | | Descriptor: | 5-[(5,6-dichloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GV2
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 13.75 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GUW
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 6.25 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
