4A0C
 
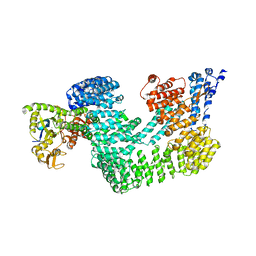 | | Structure of the CAND1-CUL4B-RBX1 complex | | Descriptor: | CULLIN-4B, CULLIN-ASSOCIATED NEDD8-DISSOCIATED PROTEIN 1, E3 UBIQUITIN-PROTEIN LIGASE RBX1, ... | | Authors: | Scrima, A, Fischer, E.S, Faty, M, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
5TUP
 
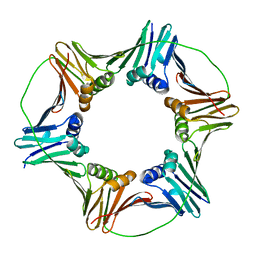 | | X-ray Crystal Structure of the Aspergillus fumigatus Sliding Clamp | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Bruning, J.B, Marshall, A.C, Kroker, A.J, Wegener, K.L, Rajapaksha, H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of the sliding clamp from the fungal pathogen Aspergillus fumigatus (AfumPCNA) and interactions with Human p21.
FEBS J., 284, 2017
|
|
6CDB
 
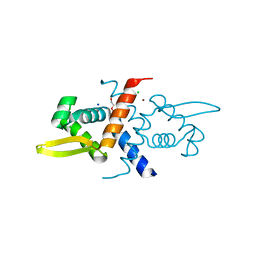 | | Crystal Structure of V66L CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, SODIUM ION, ... | | Authors: | Capdevila, D.A, Campanello, G, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
1MXJ
 
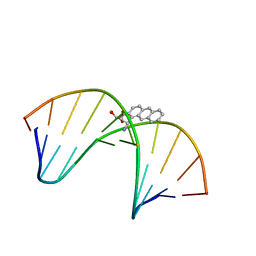 | | NMR solution structure of benz[a]anthracene-dG in ras codon 12,2; GGCAGXTGGTG | | Descriptor: | 1S,2R,3S,4R-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*AP*CP*CP*AP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*G)-3' | | Authors: | Kim, H.-Y.H, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minor Groove Orientation for the (1S,2R,3S,4R)-N2-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxy-benz[a]anthracenyl)]-2'-deoxyguanosyl Adduct in the N-ras Codon 12 sequence
Biochemistry, 42, 2003
|
|
1MNT
 
 | | SOLUTION STRUCTURE OF DIMERIC MNT REPRESSOR (1-76) | | Descriptor: | MNT REPRESSOR | | Authors: | Burgering, M.J.M, Boelens, R, Gilbert, D.E, Breg, J.N, Knight, K.L, Sauer, R.T, Kaptein, R. | | Deposit date: | 1994-06-28 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dimeric Mnt repressor (1-76).
Biochemistry, 33, 1994
|
|
1ES8
 
 | | Crystal structure of free BglII | | Descriptor: | ACETIC ACID, RESTRICTION ENDONUCLEASE BGLII | | Authors: | Lukacs, C.M, Aggarwal, A.K. | | Deposit date: | 2000-04-07 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of free BglII reveals an unprecedented scissor-like motion for opening an endonuclease.
Nat.Struct.Biol., 8, 2001
|
|
6CDA
 
 | | Crystal structure of L34A CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, GLYCEROL, ... | | Authors: | Capdevila, D.A, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
2QZ8
 
 | | Crystal structure of Mycobacterium tuberculosis Leucine response regulatory protein (LrpA) | | Descriptor: | Probable transcriptional regulatory protein | | Authors: | Manchi, C.M.R, Gokulan, K, Ioerger, T, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis LrpA, a leucine-responsive global regulator associated with starvation response.
Protein Sci., 17, 2008
|
|
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
1NFI
 
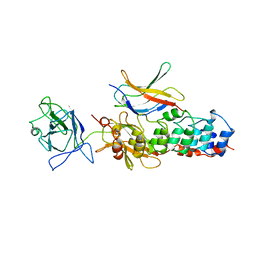 | | I-KAPPA-B-ALPHA/NF-KAPPA-B COMPLEX | | Descriptor: | I-KAPPA-B-ALPHA, NF-KAPPA-B P50, NF-KAPPA-B P65 | | Authors: | Jacobs, M.D, Harrison, S.C. | | Deposit date: | 1998-08-25 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an IkappaBalpha/NF-kappaB complex.
Cell(Cambridge,Mass.), 95, 1998
|
|
3GWS
 
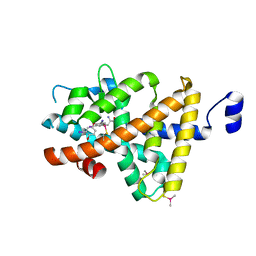 | | Crystal Structure of T3-Bound Thyroid Hormone Receptor | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor beta | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Polikarpov, I, Baxter, J.D, Webb, P. | | Deposit date: | 2009-04-01 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function
J.Mol.Biol., 360, 2006
|
|
7ZEL
 
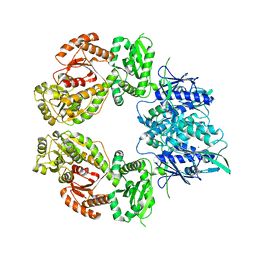 | | Human SLFN11 dimer apoenzyme | | Descriptor: | MAGNESIUM ION, Schlafen family member 11, ZINC ION | | Authors: | Metzner, F.J, Kugler, M, Wenzl, S.J, Lammens, K. | | Deposit date: | 2022-03-31 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanistic understanding of human SLFN11.
Nat Commun, 13, 2022
|
|
7ZEP
 
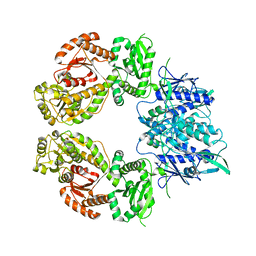 | | Human SLFN11 E209A dimer | | Descriptor: | Schlafen family member 11, ZINC ION | | Authors: | Metzner, F.J, Kugler, M, Wenzl, S.J, Lammens, K. | | Deposit date: | 2022-03-31 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic understanding of human SLFN11.
Nat Commun, 13, 2022
|
|
8V27
 
 | |
8V28
 
 | |
8V26
 
 | |
7QNQ
 
 | | Structure of the Aux2 relaxosome protein of plasmid pLS20 | | Descriptor: | Auxiliary relaxosome protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Boer, D.R, Crespo, I. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical characterization of the relaxosome auxiliary proteins encoded on the Bacillus subtilis plasmid pLS20.
Comput Struct Biotechnol J, 20, 2022
|
|
6TT5
 
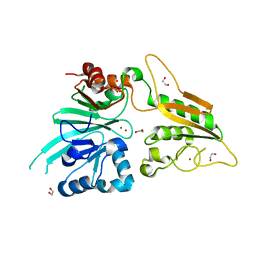 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6HN7
 
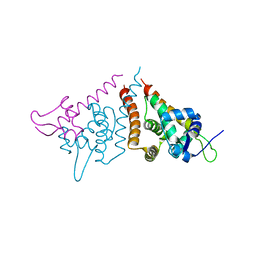 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC), Terminase small subunit | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
1TXY
 
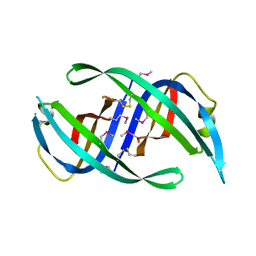 | | E. coli PriB | | Descriptor: | Primosomal replication protein n | | Authors: | Keck, J.L, Lopper, M, Holton, J.M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PriB, a component of the Escherichia coli replication restart primosome
Structure, 12, 2004
|
|
8SZR
 
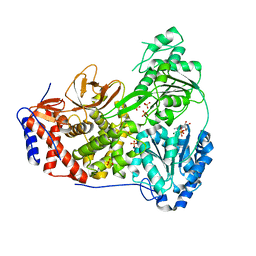 | | Dog DHX9 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RNA helicase, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SZQ
 
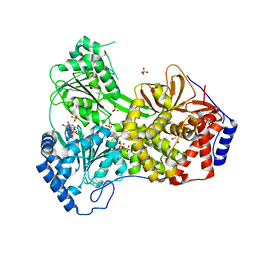 | | Cat DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7ADA
 
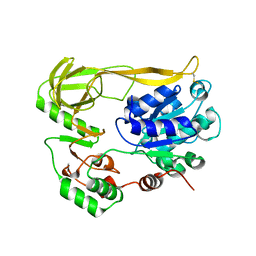 | | Crystal structure of helicase Pif1 from Thermus oshimai mutant Q164C-E409C | | Descriptor: | PIF1 helicase | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
8G6G
 
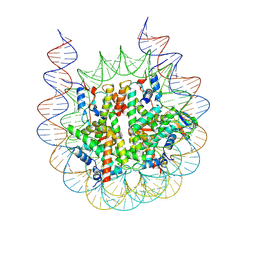 | | H2BK120ub+H3K79me2-modified nucleosome ubiquitin position 5 | | Descriptor: | 601 DNA (185-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
8TJY
 
 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
