3RF9
 
 | | X-ray structure of RlmN from Escherichia coli | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
3RLZ
 
 | |
3RNO
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with NADP. | | Descriptor: | Apolipoprotein A-I-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROM
 
 | |
3ROZ
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Nicotinamide | | Descriptor: | Apolipoprotein A-I-binding protein, NICOTINAMIDE, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RS2
 
 | | H-Ras soaked in 50% 2,2,2-trifluoroethanol: one of 10 in MSCS set | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3RS9
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P3-Di(adenosine-5') triphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSQ
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTG
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RV1
 
 | | Crystal structure of the N-terminal and RNase III domains of K. polysporus Dcr1 E224Q mutant | | Descriptor: | K. polysporus Dcr1 | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
3RY3
 
 | | Putative solute-binding protein from Yersinia pestis. | | Descriptor: | ACETATE ION, Putative solute-binding protein | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Putative solute-binding protein from Yersinia pestis.
To be Published
|
|
3RYV
 
 | | Carbonic Anhydrase complexed with N-ethyl-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, N-ethyl-4-sulfamoylbenzamide, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RUM
 
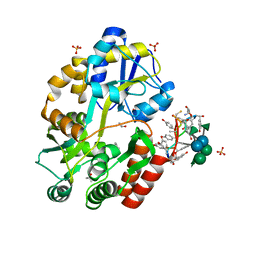 | | New strategy to analyze structures of glycopeptide antibiotic-target complexes | | Descriptor: | 3-amino-2,3,6-trideoxy-alpha-L-ribo-hexopyranose, ISOPROPYL ALCOHOL, Maltose-binding periplasmic protein, ... | | Authors: | Economou, N.J, Weeks, S.D, Grasty, K.C, Nahoum, V, Loll, P.J. | | Deposit date: | 2011-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A carrier protein strategy yields the structure of dalbavancin.
J.Am.Chem.Soc., 134, 2012
|
|
3RY1
 
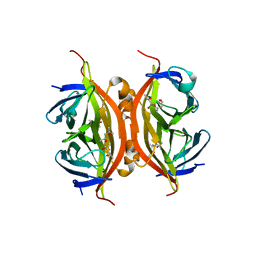 | | Wild-type core streptavidin at atomic resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | Authors: | Stenkamp, R.E, Wang, Z, Le Trong, I, Stayton, P.S, Lybrand, T.P. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Streptavidin and its biotin complex at atomic resolution.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3RYO
 
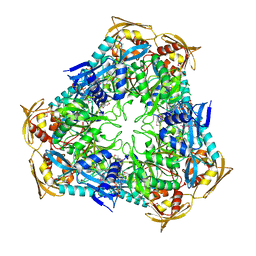 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
3RU1
 
 | |
3RVH
 
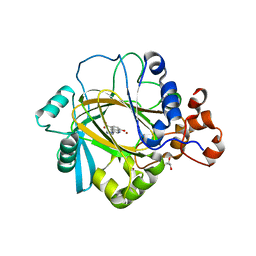 | | Crystal Structure of JMJD2A Complexed with Inhibitor | | Descriptor: | 8-hydroxy-3-(piperazin-1-yl)quinoline-5-carboxylic acid, GLYCEROL, Lysine-specific demethylase 4A, ... | | Authors: | King, O.N.F, Maloney, D.J, Tumber, A, Rai, G, Jadhav, A, Clifton, I.J, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal Structure of JMJD2A Complexed with Inhibitor
To be Published
|
|
3RWA
 
 | |
3S1B
 
 | |
3RW9
 
 | | Crystal Structure of human Spermidine Synthase in Complex with decarboxylated S-adenosylhomocysteine | | Descriptor: | 5'-S-(3-aminopropyl)-5'-thioadenosine, Spermidine synthase | | Authors: | Seckute, J, McCloskey, D.E, Thomas, H.J, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2011-05-08 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and inhibition of human spermidine synthase by decarboxylated S-adenosylhomocysteine.
Protein Sci., 20, 2011
|
|
3RWL
 
 | | Structure of P450pyr hydroxylase | | Descriptor: | Cytochrome P450 alkane hydroxylase 1 CYP153A7, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pompidor, G. | | Deposit date: | 2011-05-09 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolving P450pyr hydroxylase for highly enantioselective hydroxylation at non-activated carbon atom.
Chem.Commun.(Camb.), 48, 2012
|
|
3S28
 
 | | The crystal structure of sucrose synthase-1 in complex with a breakdown product of the UDP-glucose | | Descriptor: | 1,5-anhydro-D-arabino-hex-1-enitol, 1,5-anhydro-D-fructose, MALONIC ACID, ... | | Authors: | Zheng, Y, Garavito, R.M. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Sucrose Synthase-1 from Arabidopsis thaliana and Its Functional Implications.
J.Biol.Chem., 286, 2011
|
|
3S3Z
 
 | |
3RY9
 
 | |
