1PAG
 
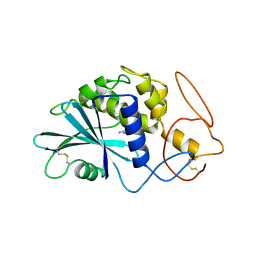 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | FORMYCIN-5'-MONOPHOSPHATE, POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
1PAH
 
 | |
1PAJ
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
1PAK
 
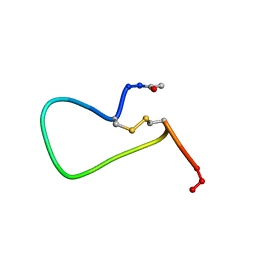 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
1PAL
 
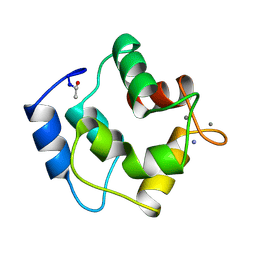 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
1PAM
 
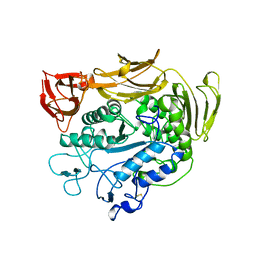 | | CYCLODEXTRIN GLUCANOTRANSFERASE | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Harata, K, Haga, K, Nakamura, A, Aoyagi, M, Yamane, K. | | Deposit date: | 1996-07-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011. Comparison of two independent molecules at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PAN
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAO PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PAO
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAO PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PAQ
 
 | |
1PAR
 
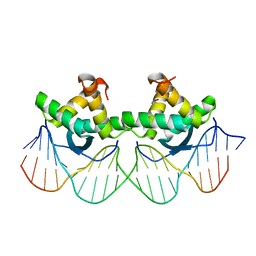 | | DNA RECOGNITION BY BETA-SHEETS IN THE ARC REPRESSOR-OPERATOR CRYSTAL STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*T P*AP*CP*TP*AP*T)- 3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*T P*AP*TP*CP*AP*T)- 3'), PROTEIN (ARC REPRESSOR) | | Authors: | Raumann, B.E, Rould, M.A, Pabo, C.O, Sauer, R.T. | | Deposit date: | 1994-03-22 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA recognition by beta-sheets in the Arc repressor-operator crystal structure.
Nature, 367, 1994
|
|
1PAU
 
 | |
1PAV
 
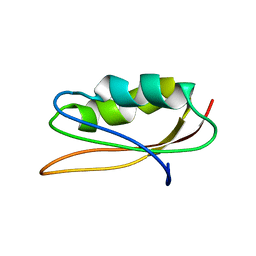 | | SOLUTION NMR STRUCTURE OF HYPOTHETICAL PROTEIN TA1414 OF THERMOPLASMA ACIDOPHILUM | | Descriptor: | Hypothetical protein Ta1170/Ta1414 | | Authors: | Monleon, D, Yee, A, Liu, C.S, Arrowsmith, C, Celda, B. | | Deposit date: | 2003-05-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein TA1414 from Thermoplasma acidophilum.
J.Biomol.Nmr, 28, 2004
|
|
1PAX
 
 | |
1PAZ
 
 | |
1PB0
 
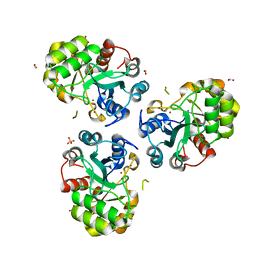 | | YCDX PROTEIN IN AUTOINHIBITED STATE | | Descriptor: | FORMIC ACID, Hypothetical protein ycdX, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Autoregulation of YcdX protein
To be Published
|
|
1PB1
 
 | |
1PB3
 
 | |
1PB5
 
 | | NMR Structure of a Prototype LNR Module from Human Notch1 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 1 | | Authors: | Vardar, D, North, C.L, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of a Prototype Lin12-Notch Repeat Module from Human Notch1
Biochemistry, 42, 2003
|
|
1PB7
 
 | |
1PB8
 
 | |
1PB9
 
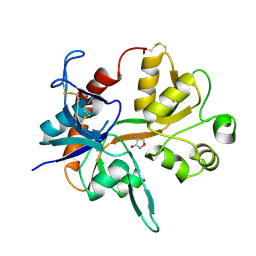 | |
1PBA
 
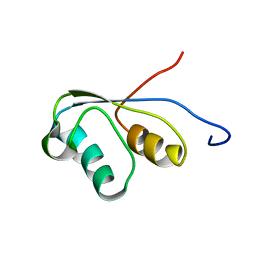 | | THE NMR STRUCTURE OF THE ACTIVATION DOMAIN ISOLATED FROM PORCINE PROCARBOXYPEPTIDASE B | | Descriptor: | PROCARBOXYPEPTIDASE B | | Authors: | Vendrell, J, Wider, G, Billeter, M, Aviles, F.X, Wuthrich, K. | | Deposit date: | 1991-11-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the activation domain isolated from porcine procarboxypeptidase B.
EMBO J., 10, 1991
|
|
1PBB
 
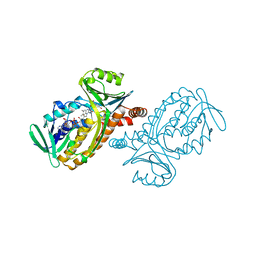 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBC
 
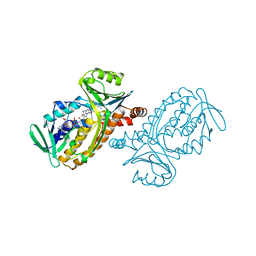 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBD
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
