8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BSC
 
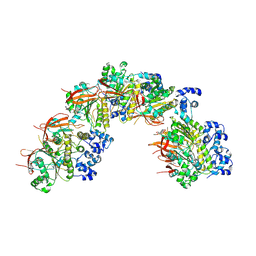 | |
8BQ2
 
 | | CryoEM structure of the pre-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
6IZI
 
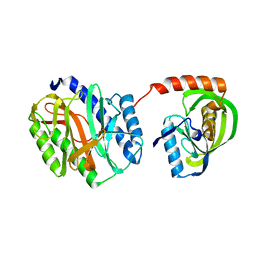 | |
6J0A
 
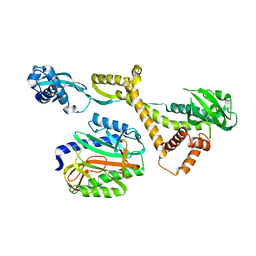 | |
6HUN
 
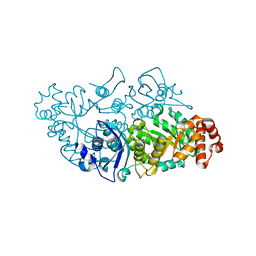 | |
8ES0
 
 | |
7NHX
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - full transcriptase (Class1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NI0
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - Replicase (class 3) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NHA
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - Endonuclease and priming loop ordered (Class2a) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NHC
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - Endonuclease ordered (Class2b) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
2N0O
 
 | |
2MWL
 
 | |
8COM
 
 | | Structure of the Nucleosome Core Particle from Trypanosoma brucei | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Burdett, H, Deak, G, Wilson, M.D. | | Deposit date: | 2023-02-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Histone divergence in trypanosomes results in unique alterations to nucleosome structure.
Nucleic Acids Res., 51, 2023
|
|
6HXW
 
 | | structure of human CD73 in complex with antibody IPH53 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, IPH53 heavy chain, ... | | Authors: | Roussel, A, Amigues, B. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Blocking Antibodies Targeting the CD39/CD73 Immunosuppressive Pathway Unleash Immune Responses in Combination Cancer Therapies.
Cell Rep, 27, 2019
|
|
8BWO
 
 | | Cryo-EM structure of nanodisc-reconstituted human MRP4 with E1202Q mutation (outward-facing occluded conformation) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, MAGNESIUM ION | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
6J9P
 
 | |
8BJF
 
 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (inward-facing conformation) | | Descriptor: | ATP-binding cassette sub-family C member 4, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWQ
 
 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with topotecan) | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWP
 
 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with methotrexate) | | Descriptor: | ATP-binding cassette sub-family C member 4, METHOTREXATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWR
 
 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with prostaglandin E2) | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
7UIO
 
 | | Mediator-PIC Early (Composite Model) | | Descriptor: | ALANINE, ASPARTIC ACID, DNA (37-MER), ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
6K0K
 
 | |
6HCY
 
 | | human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA. | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
2MYG
 
 | | Solution structure of the dithiolic glutaredoxin 2-C-Grx1 from the pathogen Trypanosoma brucei brucei | | Descriptor: | Dithiol glutaredoxin 1 | | Authors: | Sturlese, M, Stefani, M, Manta, B, Mammi, S, Comini, M, Bellanda, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dithiolic glutaredoxin 2-C-Grx1 from the pathogen Trypanosoma brucei brucei
To be Published
|
|
