7KJ4
 
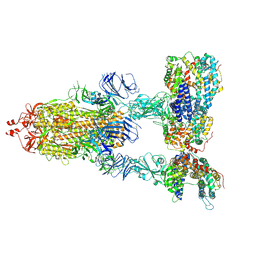 | | SARS-CoV-2 Spike Glycoprotein with three ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KMZ
 
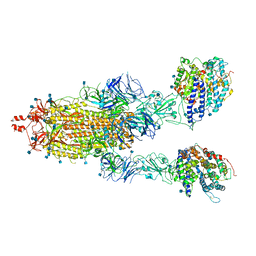 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
7LO4
 
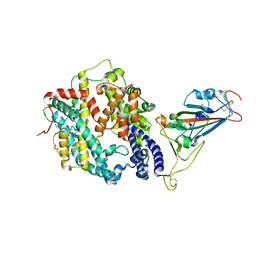 | |
7M8S
 
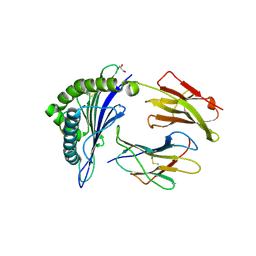 | | Crystal Structure of HLA-A*02:01 in complex with KLNDLCFTNV, an 10-mer epitope from SARS-CoV-2 Spike (S386-395) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
7M8U
 
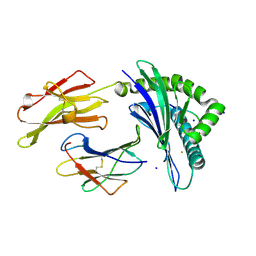 | | Crystal Structure of HLA-B*35:01 in complex with IPFAMQMAY, an 9-mer epitope from SARS-CoV-2 spike (S896-904) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen B, alpha chain, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
7M8T
 
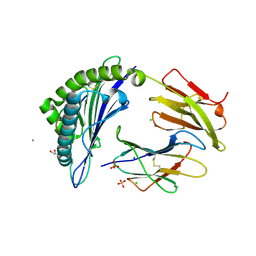 | | Crystal Structure of HLA-A*11:01 in complex with NSASFSTFK, an 9-mer epitope from SARS-CoV-2 spike (S370-378) | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MKB
 
 | |
7N6E
 
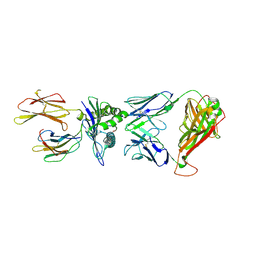 | | TCR peptide HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1, ... | | Authors: | Chaurasia, P, Rossjohn, J, Petersen, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
7N1A
 
 | | SARS-CoV-2 YLQ peptide binds to HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1B
 
 | | SARS-CoV-2 RLQ peptide binds to HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N6D
 
 | | HLA peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Chaurasia, P, Petersen, J, Rossjohn, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
7N1F
 
 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 binds to YLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1E
 
 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 binds to RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7NXC
 
 | |
7BH9
 
 | | SARS-CoV-2 RBD-62 in complex with ACE2 peptidase domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Elad, N, Dym, O, Zahradnik, J, Schreiber, G. | | Deposit date: | 2021-01-11 | | Release date: | 2021-02-17 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution.
Nat Microbiol, 6, 2021
|
|
7C8D
 
 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DQA
 
 | | Cryo-EM structure of SARS-CoV2 RBD-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, J, Lan, J, Wang, X.Q, Wang, H.W. | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reduced graphene oxide membrane as supporting film for high-resolution cryo-EM
Biophys Rep, 7, 2022
|
|
7EKE
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
