5ZFO
 
 | | NMR structure of IRD12 from Capsicum annum. | | Descriptor: | Pin-II type proteinase inhibitor 38 | | Authors: | Gartia, J, Barnwal, R.P, Chary, K.V.R. | | Deposit date: | 2018-03-06 | | Release date: | 2019-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of inhibitory repeat domain variant 12, a plant protease inhibitor from Capsicum annuum, and its structural relationship to other plant protease inhibitors.
J.Biomol.Struct.Dyn., 2019
|
|
6V1W
 
 | |
6LUL
 
 | | NMR structure and dynamics studies of yeast respiratory super-complex factor 2 in micelles | | Descriptor: | Respiratory supercomplex factor 2, mitochondrial | | Authors: | Zhou, S, Pontus, P, Peter, B, Maler, L, Adelroth, P. | | Deposit date: | 2020-01-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics Studies of Yeast Respiratory Supercomplex Factor 2.
Structure, 29, 2021
|
|
5UNK
 
 | |
6MJD
 
 | | NMR Solution structure of GIIIC | | Descriptor: | ARG-ASP-CYS-CYS-THR-HYP-HYP-LYS-LYS-CYS-LYS-ASP-ARG-ARG-CYS-LYS-HYP-LEU-LYS-CYS-CYS-ALA-NH2 | | Authors: | Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2018-09-20 | | Release date: | 2018-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of mu-Conotoxin GIIIC: Leucine 18 Induces Local Repacking of the N-Terminus Resulting in Reduced NaVChannel Potency.
Molecules, 23, 2018
|
|
4AXP
 
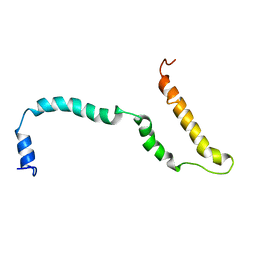 | | NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. | | Descriptor: | 12 KDA HEAT SHOCK PROTEIN | | Authors: | Herbert, A.P, Riesen, M, Bloxam, L, Kosmidou, E, Wareing, B.M, Johnson, J.R, Phelan, M.M, Pennington, S.R, Lian, L.Y, Morgan, A. | | Deposit date: | 2012-06-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Hsp12, a Protein Induced by and Required for Dietary Restriction-Induced Lifespan Extension in Yeast.
Plos One, 7, 2012
|
|
1PAV
 
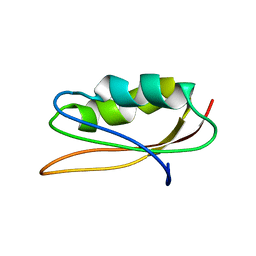 | | SOLUTION NMR STRUCTURE OF HYPOTHETICAL PROTEIN TA1414 OF THERMOPLASMA ACIDOPHILUM | | Descriptor: | Hypothetical protein Ta1170/Ta1414 | | Authors: | Monleon, D, Yee, A, Liu, C.S, Arrowsmith, C, Celda, B. | | Deposit date: | 2003-05-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein TA1414 from Thermoplasma acidophilum.
J.Biomol.Nmr, 28, 2004
|
|
1OP1
 
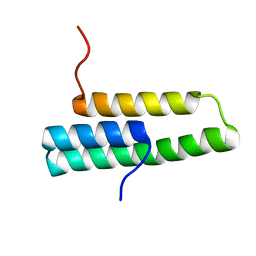 | | Solution NMR structure of domain 1 of receptor associated protein | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein precursor | | Authors: | Wu, Y, Migliorini, M, Yu, P, Strickland, D.K, Wang, Y.-X. | | Deposit date: | 2003-03-04 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of domain 1 of receptor associated protein.
J.Biomol.Nmr, 26, 2003
|
|
1NAJ
 
 | | High resolution NMR Structure Of DNA Dodecamer Determined In Aqueous Dilute Liquid Crystalline Phase | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | Wu, Z, Delaglio, F, Tjandra, N, Zhurkin, V, Bax, A. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and (31)P chemical shift anisotropy.
J.Biomol.Nmr, 26, 2003
|
|
2XI8
 
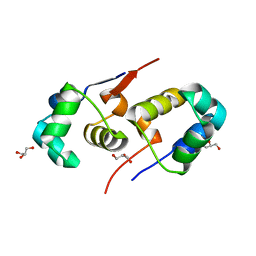 | | High resolution structure of native CylR2 | | Descriptor: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-06-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
1AW6
 
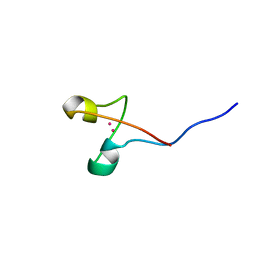 | | GAL4 (CD), NMR, 24 STRUCTURES | | Descriptor: | CADMIUM ION, GAL4 (CD) | | Authors: | Baleja, J.D, Wagner, G. | | Deposit date: | 1997-10-10 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the DNA-binding domain of GAL4 and use of 3J(113Cd,1H) in structure determination.
J.Biomol.NMR, 10, 1997
|
|
1MZT
 
 | |
1BMR
 
 | | ALPHA-LIKE TOXIN LQH III FROM SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 25 STRUCTURES | | Descriptor: | LQH III ALPHA-LIKE TOXIN | | Authors: | Krimm, I, Gilles, N, Sautiere, P, Stankiewicz, M, Pelhate, M, Gordon, D, Lancelin, J.-M. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures and activity of a novel alpha-like toxin from the scorpion Leiurus quinquestriatus hebraeus.
J.Mol.Biol., 285, 1999
|
|
1CEU
 
 | |
8DFZ
 
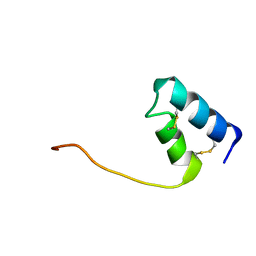 | |
1Q5F
 
 | | NMR Structure of Type IVb pilin (PilS) from Salmonella typhi | | Descriptor: | PilS | | Authors: | Xu, X.F, Tan, Y.W, Hackett, J, Zhang, M, Mok, Y.K. | | Deposit date: | 2003-08-07 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Type IVb Pilin from Salmonella typhi and Its Assembly into Pilus
J.Biol.Chem., 279, 2004
|
|
1Q9F
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX.
J.Mol.Biol., 336, 2004
|
|
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | Descriptor: | GLUTAREDOXIN 3, GLUTATHIONE | | Authors: | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-30 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
1OKD
 
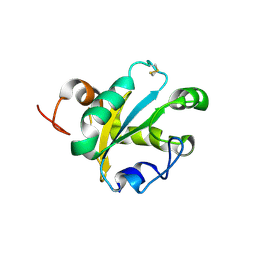 | | NMR-structure of tryparedoxin 1 | | Descriptor: | TRYPAREDOXIN 1 | | Authors: | Krumme, D, Budde, H, Hecht, H.-J, Menge, U, Ohlenschlager, O, Ross, A, Wissing, J, Wray, V, Flohe, L. | | Deposit date: | 2003-07-22 | | Release date: | 2003-08-28 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR studies of the interaction of tryparedoxin with redox-inactive substrate homologues.
Biochemistry, 42, 2003
|
|
1P6S
 
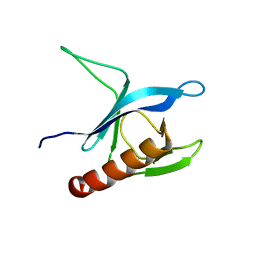 | | Solution Structure of the Pleckstrin Homology Domain of Human Protein Kinase B beta (Pkb/Akt) | | Descriptor: | RAC-beta serine/threonine protein kinase | | Authors: | Auguin, D, Barthe, P, Auge-Senegas, M.T, Stern, M.H, Noguchi, M, Roumestand, C. | | Deposit date: | 2003-04-30 | | Release date: | 2004-05-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the pleckstrin homology domain of the human
protein kinase B (PKB/Akt). Interaction with inositol phosphates.
J.BIOMOL.NMR, 28, 2004
|
|
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
1DBY
 
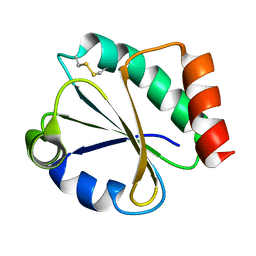 | | NMR STRUCTURES OF CHLOROPLAST THIOREDOXIN M CH2 FROM THE GREEN ALGA CHLAMYDOMONAS REINHARDTII | | Descriptor: | CHLOROPLAST THIOREDOXIN M CH2 | | Authors: | Lancelin, J.-M, Guilhaudis, L, Krimm, I, Blackledge, M.J, Marion, D. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thioredoxin m from the green alga Chlamydomonas reinhardtii.
Proteins, 41, 2000
|
|
1OW9
 
 | | NMR Structure of the Active Conformation of the VS Ribozyme Cleavage Site | | Descriptor: | A mimic of the VS Ribozyme Hairpin Substrate | | Authors: | Hoffmann, B, Mitchell, G.T, Gendron, P, Major, F, Andersen, A.A, Collins, R.A, Legault, P. | | Deposit date: | 2003-03-28 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Active Conformation of the Varkud satellite Ribozyme Cleavage Site
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NYA
 
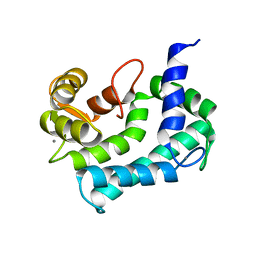 | | NMR SOLUTION STRUCTURE OF CALERYTHRIN, AN EF-HAND CALCIUM-BINDING PROTEIN | | Descriptor: | CALCIUM ION, Calerythrin | | Authors: | Tossavainen, H, Permi, P, Annila, A, Kilpelainen, I, Drakenberg, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of calerythrin, an EF-hand calcium-binding protein from Saccharopolyspora erythraea
Eur.J.Biochem., 270, 2003
|
|
1MQZ
 
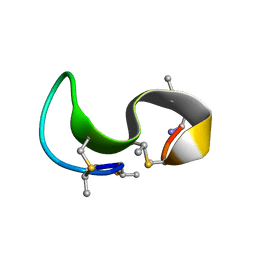 | | NMR solution structure of type-B lantibiotics mersacidin bound to lipid II in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
