1XWA
 
 | | Drospohila thioredoxin, oxidized, P41212 | | Descriptor: | CADMIUM ION, CHLORIDE ION, thioredoxin | | Authors: | Wahl, M.C, Irmler, A, Hecker, B, Schirmer, R.H, Becker, K. | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative structural analysis of oxidized and reduced thioredoxin from Drosophila melanogaster
J.Mol.Biol., 345, 2005
|
|
1XW9
 
 | | Drosophila thioredoxin, oxidized, P21 | | Descriptor: | thioredoxin | | Authors: | Wahl, M.C, Irmler, A, Hecker, B, Schirmer, R.H, Becker, K. | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparative structural analysis of oxidized and reduced thioredoxin from Drosophila melanogaster
J.Mol.Biol., 345, 2005
|
|
1ZJI
 
 | | Aquifex aeolicus KDO8PS R106G mutant in complex with 2PGA and R5P | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop
Biochemistry, 44, 2005
|
|
1YXA
 
 | | Serpina3n, a murine orthologue of human antichymotrypsin | | Descriptor: | serine (or cysteine) proteinase inhibitor, clade A, member 3N | | Authors: | Horvath, A.J, Irving, J.A, Law, R.H, Rossjohn, J, Bottomley, S.P, Quinsey, N.S, Pike, R.N, Coughlin, P.B, Whisstock, J.C. | | Deposit date: | 2005-02-20 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins.
J.Biol.Chem., 280, 2005
|
|
1Z47
 
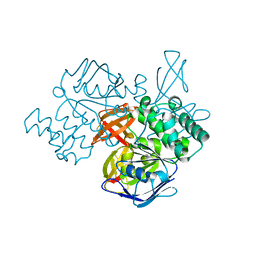 | | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius | | Descriptor: | CHLORIDE ION, putative ABC-transporter ATP-binding protein | | Authors: | Scheffel, F, Demmer, U, Warkentin, E, Huelsmann, A, Schneider, E, Ermler, U. | | Deposit date: | 2005-03-15 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius
Febs Lett., 579, 2005
|
|
3HYV
 
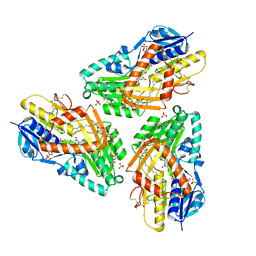 | | 3-D X-Ray structure of the sulfide:quinone oxidoreductase from the hyperthermophilic bacterium Aquifex aeolicus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marcia, M, Ermler, U, Peng, G.H, Michel, H. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1Y60
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 with bound 5,10-methylene tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Formaldehyde-activating enzyme fae | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
3HYX
 
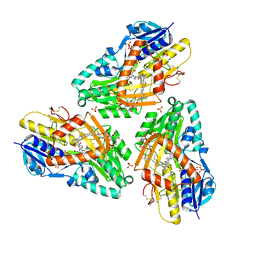 | | 3-D X-Ray structure of the sulfide:quinone oxidoreductase from Aquifex aeolicus in complex with Aurachin C | | Descriptor: | 1-hydroxy-2-methyl-3-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]quinolin-4(1H)-one, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marcia, M, Ermler, U, Peng, G.H, Michel, H. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1ZHA
 
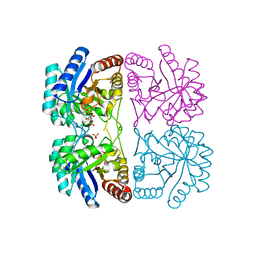 | | A. aeolicus KDO8PS R106G mutant in complex with PEP and R5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop.
Biochemistry, 44, 2005
|
|
1Z69
 
 | | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Ermler, U, Hagemeier, C.H, Thauer, R.K, Shima, S. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family
Protein Sci., 14, 2005
|
|
3H65
 
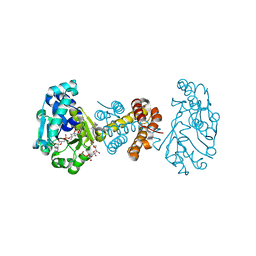 | | The Crystal Structure of C176A Mutated [Fe]-Hydrogenase (Hmd) Holoenzyme in Complex with Methylenetetrahydromethanopterin | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, ... | | Authors: | Hiromoto, T, Warkentin, E, Shima, S, Ermler, U. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of an [Fe]-hydrogenase-substrate complex reveals the framework for H2 activation.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1XWC
 
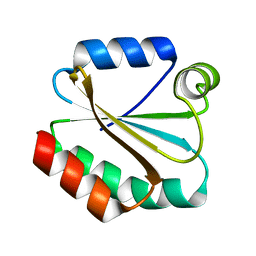 | | Drosophila thioredoxin, reduced, P6522 | | Descriptor: | thioredoxin | | Authors: | Wahl, M.C, Irmler, A, Hecker, B, Schirmer, R.H, Becker, K. | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparative structural analysis of oxidized and reduced thioredoxin from Drosophila melanogaster
J.Mol.Biol., 345, 2005
|
|
1XWB
 
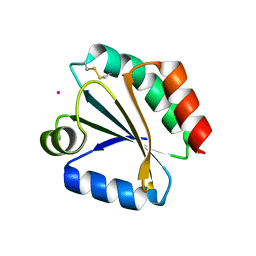 | | Drospohila thioredoxin, oxidized, P42212 | | Descriptor: | CADMIUM ION, thioredoxin | | Authors: | Wahl, M.C, Irmler, A, Hecker, B, Schirmer, R.H, Becker, K. | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative structural analysis of oxidized and reduced thioredoxin from Drosophila melanogaster
J.Mol.Biol., 345, 2005
|
|
2A5W
 
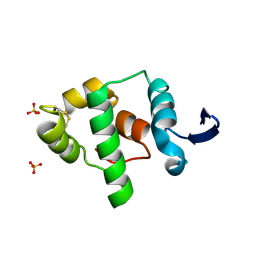 | | Crystal structure of the oxidized gamma-subunit of the dissimilatory sulfite reductase (DsrC) from Archaeoglobus fulgidus | | Descriptor: | SULFATE ION, sulfite reductase, desulfoviridin-type subunit gamma (dsvC) | | Authors: | Mander, G.J, Weiss, M.S, Hedderich, R, Kahnt, J, Ermler, U, Warkentin, E. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of the gamma-subunit of a dissimilatory sulfite reductase: Fixed and flexible C-terminal arms.
Febs Lett., 579, 2005
|
|
1Y5Y
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 | | Descriptor: | CALCIUM ION, Formaldehyde-activating enzyme fae, SODIUM ION | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
3MM7
 
 | | Dissimilatory sulfite reductase carbon monoxide complex | | Descriptor: | CARBON MONOXIDE, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3MMC
 
 | | Structure of the dissimilatory sulfite reductase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Schiffer, A, Parey, K, Warkentin, E, Diederichs, K, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the dissimilatory sulfite reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Mol.Biol., 379, 2008
|
|
3MM9
 
 | | Dissimilatory sulfite reductase nitrite complex | | Descriptor: | IRON/SULFUR CLUSTER, NITRITE ION, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3MPJ
 
 | | Structure of the glutaryl-coenzyme A dehydrogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
3M97
 
 | | Structure of the soluble domain of cytochrome c552 with its flexible linker segment from Paracoccus denitrificans | | Descriptor: | Cytochrome c-552, HEME C, ZINC ION | | Authors: | Rajendran, C, Ermler, U, Ludwig, B, Michel, H. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structure at 1.5 A resolution of cytochrome c(552) with its flexible linker segment, a membrane-anchored protein from Paracoccus denitrificans.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MM8
 
 | | Dissimilatory sulfite reductase nitrate complex | | Descriptor: | IRON/SULFUR CLUSTER, NITRATE ION, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3MM5
 
 | | Dissimilatory sulfite reductase in complex with the substrate sulfite | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3MPI
 
 | | Structure of the glutaryl-coenzyme A dehydrogenase glutaryl-CoA complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, glutaryl-coenzyme A | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
3MK7
 
 | | The structure of CBB3 cytochrome oxidase | | Descriptor: | 30-mer peptide, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Buschmann, S, Warkentin, E, Michel, H, Ermler, U. | | Deposit date: | 2010-04-14 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of cbb3 Cytochrome Oxidase Provides Insights into Proton Pumping
Science, 329, 2010
|
|
3MM6
 
 | | Dissimilatory sulfite reductase cyanide complex | | Descriptor: | CYANIDE ION, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
