2Q63
 
 | | HIV-1 PR mutant in complex with nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, PROTEASE RETROPEPSIN | | Authors: | Rezacova, P, Kozisek, M, Saskova, K, Brynda, J, Konvalinka, J. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants
J.Mol.Biol., 374, 2007
|
|
2PYM
 
 | | HIV-1 PR mutant in complex with nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, PROTEASE RETROPEPSIN | | Authors: | Rezacova, P, Kozisek, M, Saskova, K, Brynda, J, Konvalinka, J. | | Deposit date: | 2007-05-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants
J.Mol.Biol., 374, 2007
|
|
2Q64
 
 | | HIV-1 PR mutant in complex with nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, PROTEASE RETROPEPSIN | | Authors: | Rezacova, P, Kozisek, M, Saskova, K, Brynda, J, Konvalinka, J. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants
J.Mol.Biol., 374, 2007
|
|
4BLL
 
 | | Crystal Structure of Fungal Versatile Peroxidase I from Pleurotus ostreatus - Crystal Form II | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE I, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
2Y5P
 
 | |
4OKN
 
 | | Crystal structure of human muscle L-lactate dehydrogenase, ternary complex with NADH and oxalate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, KANAMYCIN A, L-lactate dehydrogenase A chain, ... | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4BLK
 
 | | Crystal Structure of Fungal Versatile Peroxidase I from Pleurotus ostreatus - Crystal Form I | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE I | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.049 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
1PNH
 
 | | SOLUTION STRUCTURE OF PO5-NH2, A SCORPION TOXIN ANALOG WITH HIGH AFFINITY FOR THE APAMIN-SENSITIVE POTASSIUM CHANNEL | | Descriptor: | SCORPION TOXIN | | Authors: | Meunier, S, Bernassau, J.-M, Sabatier, J.-M, Martin-Eauclaire, M.-F, Van Rietschoten, J, Cambillau, C, Darbon, H. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P05-NH2, a scorpion toxin analog with high affinity for the apamin-sensitive potassium channel.
Biochemistry, 32, 1993
|
|
4CFS
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE | | Descriptor: | 1-H-3-HYDROXY-4-OXOQUINALDINE 2,4-DIOXYGENASE, 3-HYDROXY-2-METHYLQUINOLIN-4(1H)-ONE, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Origin of the Proton-Transfer Step in the Cofactor-Free 1-H-3-Hydroxy-4-Oxoquinaldine 2,4- Dioxygenase: Effect of the Basicity of an Active Site His Residue.
J.Biol.Chem., 289, 2014
|
|
4BLY
 
 | | CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM V | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
3RB5
 
 | | Crystal structure of calcium binding domain CBD12 of CALX1.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Na/Ca exchange protein, ... | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2011-03-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Ca(2+) Inhibitory Mechanism of Drosophila Na(+)/Ca(2+) Exchanger CALX and Its Modification by Alternative Splicing.
Structure, 19, 2011
|
|
6V69
 
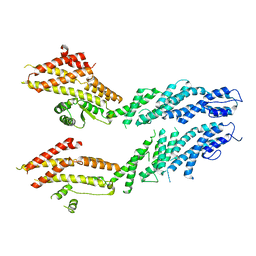 | | Structures of GCP4 and GCP5 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 4, Gamma-tubulin complex component 5 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V6C
 
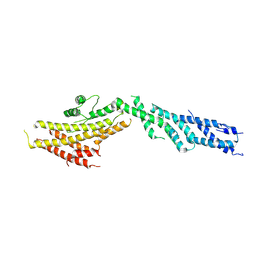 | | Structure of GCP6 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 6 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
2WU6
 
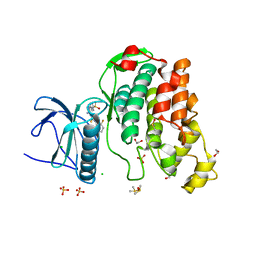 | | Crystal Structure of the Human CLK3 in complex with DKI | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CHLORIDE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Philips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
6USY
 
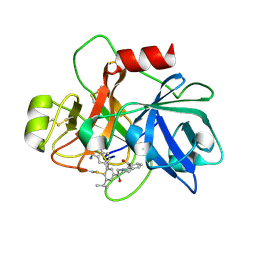 | | COAGULATION FACTOR XI CATALYTIC DOMAIN (C123S) IN COMPLEX WITH NVP-XIV936 | | Descriptor: | 1-[(2S)-2-{3-[(3S)-3-amino-2,3-dihydro-1-benzofuran-5-yl]-5-(propan-2-yl)phenyl}-2-hydroxyethyl]-1H-indole-7-carboxylic acid, Coagulation factor XIa light chain | | Authors: | Weihofen, W.A, Clark, K, Nunes, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
2WU7
 
 | | Crystal Structure of the Human CLK3 in complex with V25 | | Descriptor: | CHLORIDE ION, DUAL SPECIFICITY PROTEIN KINASE CLK3, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Phillips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Bracher, F, Huber, K, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
7ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C1A COMPLEX WITH CYANIDE AND FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, CYANIDE ION, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-26 | | Release date: | 2000-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
3NSU
 
 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
3TGR
 
 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7AVC
 
 | | DoBi scaffold based on PIH1D1 N-terminal domain | | Descriptor: | GLYCEROL, PIH1 domain-containing protein 1, SODIUM ION | | Authors: | Kolenko, P, Pham, N.P, Pavlicek, J, Mikulecky, P, Schneider, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Binder (ProBi) as a New Class of Structurally Robust Non-Antibody Protein Scaffold for Directed Evolution.
Viruses, 13, 2021
|
|
7KH2
 
 | | Structure of N-citrylornithine decarboxylase bound with PLP | | Descriptor: | GLYCEROL, N-citrylornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M, Michael, A. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-16 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Alternative pathways utilize or circumvent putrescine for biosynthesis of putrescine-containing rhizoferrin.
J.Biol.Chem., 296, 2020
|
|
6V6S
 
 | | Structure of the native human gamma-tubulin ring complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Gamma-tubulin complex component 2, ... | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
3IJE
 
 | | Crystal structure of the complete integrin alhaVbeta3 ectodomain plus an Alpha/beta transmembrane fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xiong, J.-P, Mahalingham, B, Rui, X, Hyman, B.T, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment.
J.Cell Biol., 186, 2009
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6V5V
 
 | | Structure of gamma-tubulin in the native human gamma-tubulin ring complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Tubulin gamma-1 chain | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
