1HX6
 
 | | P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAJOR CAPSID PROTEIN, ... | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2001-01-11 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6BT3
 
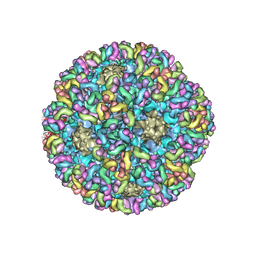 | | High-Resolution Structure Analysis of Antibody V5 Conformational Epitope on Human Papillomavirus 16 | | Descriptor: | Major capsid protein L1, V5 Fab Heavy-chain, V5 Fab Light-chain | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2017-12-05 | | Release date: | 2018-01-17 | | Last modified: | 2018-01-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16.
Viruses, 9, 2017
|
|
2CQS
 
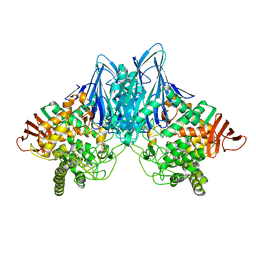 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate | | Descriptor: | Cellobiose Phosphorylase, SULFATE ION, beta-D-glucopyranose | | Authors: | Hidaka, M, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-05-20 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural dissection of the reaction mechanism of cellobiose phosphorylase.
Biochem.J., 398, 2006
|
|
1LQD
 
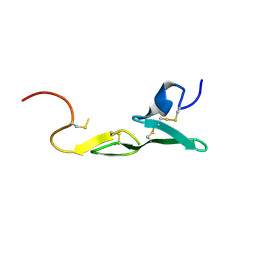 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 45. | | Descriptor: | 1-(3-CARBAMIMIDOYL-BENZYL)-4-METHYL-1H-INDOLE-2-CARBOXYLIC ACID 3,5-DIMETHYL-BENZYLAMIDE, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Loenze, P, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-10 | | Release date: | 2003-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Med.Chem., 45, 2002
|
|
1LQT
 
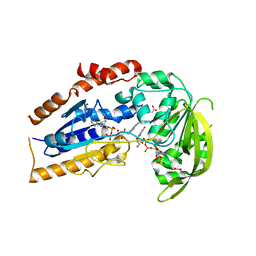 | | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bossi, R.T, Aliverti, A, Raimondi, D, Fischer, F, Zanetti, G, Ferrari, D, Tahallah, N, Maier, C.S, Heck, A.J.R, Rizzi, M, Mattevi, A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-13 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase.
Biochemistry, 41, 2002
|
|
6CN6
 
 | | RORC2 LBD complexed with compound 34 | | Descriptor: | 3-cyano-N-{3-[1-(cyclopentanecarbonyl)piperidin-4-yl]-1,4-dimethyl-1H-indol-5-yl}benzamide, Nuclear receptor ROR-gamma | | Authors: | Kauppi, B, Vajdos, F. | | Deposit date: | 2018-03-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 3-Cyano- N-(3-(1-isobutyrylpiperidin-4-yl)-1-methyl-4-(trifluoromethyl)-1 H-pyrrolo[2,3- b]pyridin-5-yl)benzamide: A Potent, Selective, and Orally Bioavailable Retinoic Acid Receptor-Related Orphan Receptor C2 Inverse Agonist.
J. Med. Chem., 61, 2018
|
|
1LGV
 
 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 100K | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | Deposit date: | 2002-04-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
1LKV
 
 | |
5H1W
 
 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Mn2+ and L(+)-Erythrulose | | Descriptor: | L-Erythrulose, MANGANESE (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-10-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
1L1Y
 
 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1JKT
 
 | | TETRAGONAL CRYSTAL FORM OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-13 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1JXK
 
 | | Role of ethe mobile loop in the mehanism of human salivary amylase | | Descriptor: | Alpha-amylase, salivary, CALCIUM ION, ... | | Authors: | Ramasubbu, N, Ragunath, C, Wang, Z. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of a mobile loop in substrate binding and enzyme activity of human salivary amylase
J.Mol.Biol., 325, 2003
|
|
1JJA
 
 | |
1JMO
 
 | | Crystal Structure of the Heparin Cofactor II-S195A Thrombin Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Baglin, T.P, Carrell, R.W, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2001-07-19 | | Release date: | 2002-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K2V
 
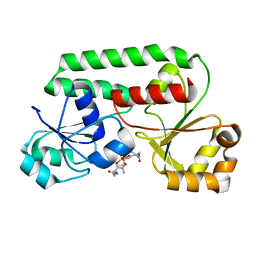 | | E. COLI PERIPLASMIC PROTEIN FHUD COMPLEXED WITH DESFERAL | | Descriptor: | DEFEROXAMINE MESYLATE FE(III) COMPLEX, Ferrichrome-binding periplasmic protein | | Authors: | Clarke, T.E, Braun, V, Winkelmann, G, Tari, L.W, Vogel, H.J. | | Deposit date: | 2001-09-29 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin.
J.Biol.Chem., 277, 2002
|
|
2ENB
 
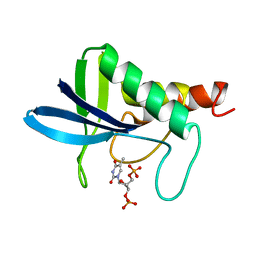 | |
1JVM
 
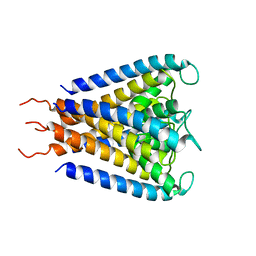 | |
1JIQ
 
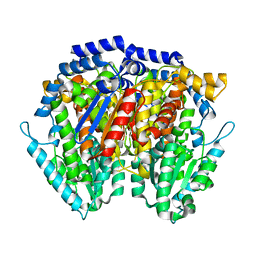 | | Crystal Structure of Human Autocrine Motility Factor | | Descriptor: | autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
1JYI
 
 | | CONCANAVALIN A/12-MER PEPTIDE COMPLEX | | Descriptor: | 12-mer peptide, CALCIUM ION, Concanavalin-Br, ... | | Authors: | Jain, D, Kaur, K.J, Sundaravadivel, B, Salunke, D.M. | | Deposit date: | 2001-09-12 | | Release date: | 2002-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Functional Consequences of Peptide-carbohydrate Mimicry. Crystal Structure of a Carbohydrate-mimicking Peptide Bound to Concanavalin A.
J.Biol.Chem., 275, 2000
|
|
7JGT
 
 | | Crystal Structure of FN3tt | | Descriptor: | Fibronectin type-III domain-containing protein | | Authors: | Luo, J, Malia, T.J. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
1LPU
 
 | | Low Temperature Crystal Structure of the Apo-form of the catalytic subunit of protein kinase CK2 from Zea mays | | Descriptor: | BENZAMIDINE, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
1LI0
 
 | | Crystal structure of TEM-32 beta-Lactamase at 1.6 Angstrom | | Descriptor: | BICARBONATE ION, Class A beta-Lactamase- TEM-32, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
1L3D
 
 | |
1L4J
 
 | | Holliday Junction TCGGTACCGA with Na and Ca Binding Sites. | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', CALCIUM ION, SODIUM ION | | Authors: | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | Deposit date: | 2002-03-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational and Hydration Effects of Site-selective Sodium, Calcium and
Strontium Ion Binding to the DNA Holliday Junction Structure
d(TCGGTACCGA)4
J.Mol.Biol., 327, 2003
|
|
1L2K
 
 | | Neutron Structure Determination of Sperm Whale Met-Myoglobin at 1.5A Resolution. | | Descriptor: | AMMONIUM CATION WITH D, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Tanaka, I, Engler, N, Niimura, N, Parak, F.G. | | Deposit date: | 2002-02-21 | | Release date: | 2002-08-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Hydrogen and deuterium in myoglobin as seen by a neutron structure determination at 1.5 A resolution.
Biophys.Chem., 95, 2002
|
|
