7U4Z
 
 | |
7U8F
 
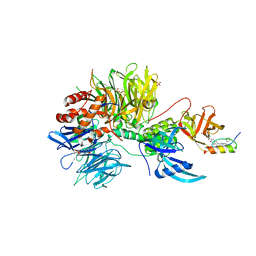 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA damage-binding protein 1, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
7P0N
 
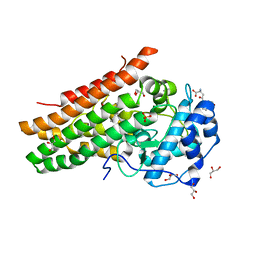 | | Crystal structure of L-Trp/Indoleamine 2,3-dioxygenagse 1 (hIDO1) complex with the JK-loop refined in the open conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Mirgaux, M, Wouters, J. | | Deposit date: | 2021-06-30 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Temporary Intermediates of L-Trp Along the Reaction Pathway of Human Indoleamine 2,3-Dioxygenase 1 and Identification of an Exo Site.
Int J Tryptophan Res, 14, 2021
|
|
7TZM
 
 | |
7TZY
 
 | |
7U00
 
 | |
7TRT
 
 | |
7TRU
 
 | |
7TZN
 
 | |
7TZW
 
 | |
7P6L
 
 | | Heme domain of CYP505A30, a fungal hydroxylase from Myceliophthora thermophila, bound to dodecanoic acid | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, LAURIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Opperman, D.J, Aschenbrenner, J.C, Tolmie, C, Ebrecht, A.C. | | Deposit date: | 2021-07-16 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of the fungal hydroxylase, CYP505A30, and rational transfer of mutation data from CYP102A1 to alter regioselectivity
Catalysis Science And Technology, 11, 2021
|
|
7TZX
 
 | | The crystal structure of WT CYP199A4 bound to 4-chloromethylbenzoic acid | | Descriptor: | 4-(chloromethyl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Scaffidi-Muta, J, Bell, S.G. | | Deposit date: | 2022-02-16 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.414 Å) | | Cite: | Cytochrome P450-catalyzed oxidation of halogen-containing substrates.
J.Inorg.Biochem., 244, 2023
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
8WZH
 
 | |
3HPK
 
 | |
7THG
 
 | |
7TNV
 
 | |
8WZM
 
 | |
8WZJ
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WZK
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WTK
 
 | | Crystal structure of Cas3-DExD+MG | | Descriptor: | CRISPR-associated helicase Cas3, MAGNESIUM ION | | Authors: | Mo, X. | | Deposit date: | 2023-10-18 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural insight into dynamic characteristic of the polymerized forms of kinetoplastid membrane protein-11
To be published
|
|
7SBJ
 
 | |
7RTN
 
 | | Cryo-EM structure of bluetongue virus capsid protein VP5 at low endosomal pH | | Descriptor: | Outer capsid protein VP5 | | Authors: | Xia, X, Wu, W.N, Cui, Y.X, Roy, P, Zhou, Z.H. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bluetongue virus capsid protein VP5 perforates membranes at low endosomal pH during viral entry.
Nat Microbiol, 6, 2021
|
|
7OQH
 
 | | CryoEM structure of the transcription termination factor Rho from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Saridakis, E, Vishwakarma, R, Lai Kee Him, J, Martin, K, Simon, I, Cohen-Gonsaud, M, Coste, F, Bron, P, Margeat, E, Boudvillain, M. | | Deposit date: | 2021-06-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structure of transcription termination factor Rho from Mycobacterium tuberculosis reveals bicyclomycin resistance mechanism.
Commun Biol, 5, 2022
|
|
7RXV
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine, Malonate (MLA) and MgF3 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, GLYCEROL, ... | | Authors: | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
