1E9W
 
 | | Structure of the macrocycle thiostrepton solved using the anomalous dispersive contribution from sulfur | | Descriptor: | THIOSTREPTON | | Authors: | Bond, C.S, Shaw, M.P, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2000-10-27 | | Release date: | 2001-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the Macrocycle Thiostrepton Solved Using the Anomalous Dispersive Contribution from Sulfur
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5OXC
 
 | | Structure of Cerulean Fluorescent Protein at 1.02 Angstrom resolution | | Descriptor: | Green fluorescent protein | | Authors: | Gotthard, G, von Stetten, D, Clavel, D, Noirclerc-Savoye, M, Royant, A. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Chromophore Isomer Stabilization Is Critical to the Efficient Fluorescence of Cyan Fluorescent Proteins.
Biochemistry, 56, 2017
|
|
5AI3
 
 | |
6R3N
 
 | |
5B27
 
 | | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid.
To Be Published
|
|
3NU3
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1S0Q
 
 | |
7A2S
 
 | |
1P7W
 
 | | Crystal structure of the complex of Proteinase K with a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Perbandt, M, Chandra, V, Banumathi, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the complex of Proteinase K with heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution
To be published
|
|
5SSZ
 
 | |
5SSX
 
 | |
6SDJ
 
 | | Human Carbonic Anhydrase II in complex with (R)-1-aminopropan-2-ol | | Descriptor: | (2~{R})-1-azanylpropan-2-ol, (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-07-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6SAC
 
 | | N-terminal expression tag remainder of human Carbonic Anhydrase II covalently modified by fragment | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 1-(1,3-dimethyl-1H-pyrazol-5-yl)methanamine, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-07-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
8RNY
 
 | | Hen Egg White Lysozyme soaked with with [H2Ind][trans-RuCl4(DMSO)(HInd)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
7T5U
 
 | | Structure of E. coli MS115-1 CapH N-terminal domain | | Descriptor: | Helix-turn-helix domain-containing protein, SULFATE ION | | Authors: | Lau, R.K, Corbett, K.D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A conserved signaling pathway activates bacterial CBASS immune signaling in response to DNA damage.
Embo J., 41, 2022
|
|
3A0M
 
 | | Structure of (PPG)4-OVG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-21 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Stabilization of triple-helical structures of collagen peptides containing a Hyp-Thr-Gly, Hyp-Val-Gly, or Hyp-Ser-Gly sequence.
Biopolymers, 95, 2011
|
|
6JGU
 
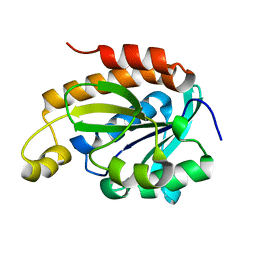 | |
6Y4E
 
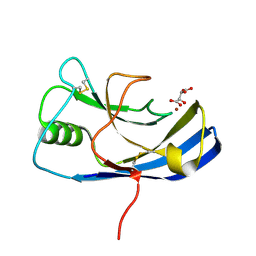 | | X-ray structure of the Zn-dependent receptor-binding domain of Proteus mirabilis MR/P fimbrial adhesin MrpH | | Descriptor: | Fimbrial adhesin, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Knight, S.D, Ubhayasekera, W, Jiang, W. | | Deposit date: | 2020-02-20 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | MrpH, a new class of metal-binding adhesin, requires zinc to mediate biofilm formation.
Plos Pathog., 16, 2020
|
|
6ETP
 
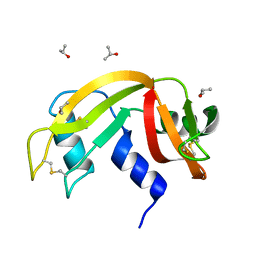 | |
6YQM
 
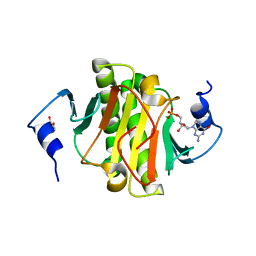 | | Human histidine triad nucleotide-binding protein 1 (hHINT1) complexed with dGMP and refined to 1.02 A | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.D, Seda, A, Nawrot, B.C. | | Deposit date: | 2020-04-17 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
4YU1
 
 | |
4CZ5
 
 | |
8GL0
 
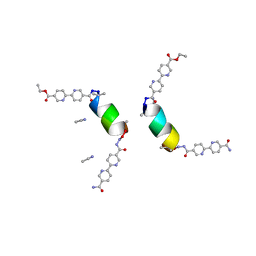 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, LEU-AIB-ALA-CYS-LEU-CYS-GLN-AIB-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GL5
 
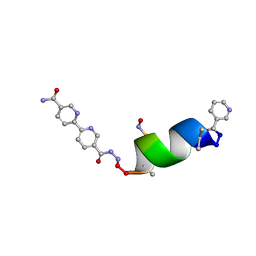 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-SER-LEU-ALA-SNC-AIB-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
