3PEO
 
 | | Crystal structure of acetylcholine binding protein complexed with metocurine | | Descriptor: | 6,6',7',12'-tetramethoxy-2,2,2',2'-tetramethyltubocuraran-2,2'-diium, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Harel, M, Yamauchi, G.J, Radic, Z, Hansen, S, Huxford, T, Taylor, P.W. | | Deposit date: | 2010-10-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The curare alkaloids: analyzing the poses of complexes with the acetylcholine binding protein in relation to structure and binding energetics
To be Published
|
|
3PE0
 
 | |
3RSV
 
 | |
4L57
 
 | | High resolutin structure of human cytosolic 5'(3')-deoxyribonucleotidase | | Descriptor: | 5'(3')-deoxyribonucleotidase, cytosolic type, GLYCEROL, ... | | Authors: | Pachl, P, Brynda, J, Rezacova, P. | | Deposit date: | 2013-06-10 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structures of human cytosolic and mitochondrial nucleotidases: implications for structure-based design of selective inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 70, 2014
|
|
4L5N
 
 | | Crystallographic Structure of HHV-1 Uracil-DNA Glycosylase complexed with the Bacillus phage PZA inhibitor protein p56 | | Descriptor: | ACETATE ION, Early protein GP1B, Uracil-DNA glycosylase | | Authors: | Cole, A.R, Sapir, O, Ryzhenkova, K, Baltulionis, G, Hornyak, P, Savva, R. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Architecturally diverse proteins converge on an analogous mechanism to inactivate Uracil-DNA glycosylase.
Nucleic Acids Res., 41, 2013
|
|
3TCR
 
 | |
3RTH
 
 | |
4FPB
 
 | |
4FBN
 
 | | Insights into structural integration of the PLCgamma regulatory region and mechanism of autoinhibition and activation based on key roles of SH2 domains | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
3RFV
 
 | | Crystal structure of Uronate dehydrogenase from Agrobacterium tumefaciens complexed with NADH and product | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-galactaro-1,5-lactone, PHOSPHATE ION, ... | | Authors: | Parkkinen, T, Rouvinen, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Uronate Dehydrogenase from Agrobacterium tumefaciens.
J.Biol.Chem., 286, 2011
|
|
3QXT
 
 | |
3QXE
 
 | | Crystal Structure of Co-type Nitrile Hydratase from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QUZ
 
 | | Structure of the mouse CD1d-NU-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
4EXA
 
 | | Crystal structure of the PA4992, the putative aldo-keto reductase from Pseudomona aeruginosa | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3QYH
 
 | | Crystal Structure of Co-type Nitrile Hydratase beta-H71L from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
4EFI
 
 | | Crystal Structure of 3-oxoacyl-(Acyl-carrier protein) Synthase from Burkholderia Xenovorans LB400 | | Descriptor: | 3-oxoacyl-(Acyl-carrier protein) synthase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Craig, T.K, Abendroth, J, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
3QUY
 
 | | Structure of the mouse CD1d-BnNH-GSL-1'-iNKT TCR complex | | Descriptor: | (2S,3R,4S,5R,6S)-6-[(2S,3S,4R)-2-(hexacosanoylamino)-3,4-dihydroxy-octadecoxy]-3,4,5-trihydroxy-N-(phenylmethyl)oxane-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
4ENZ
 
 | | Structure of human ceruloplasmin at 2.6 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Samygina, V.R, Sokolov, A.V, Bourenkov, G, Vasilyev, V.B, Bartunik, H. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins.
Plos One, 8, 2013
|
|
3NWX
 
 | | X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-12 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4EUI
 
 | | Crystal Structure of MIF L46F mutant | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Ashrafi, A, Pojer, F, Lashuel, H. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket.
Plos One, 7, 2012
|
|
4KVO
 
 | |
4KVM
 
 | |
3UU4
 
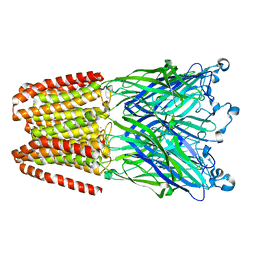 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in the crystal in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3O8M
 
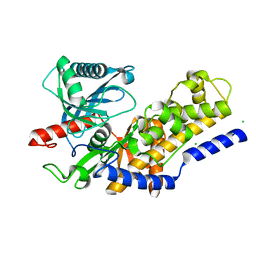 | | Crystal structure of monomeric KlHxk1 in crystal form XI with glucose bound (closed state) | | Descriptor: | CHLORIDE ION, Hexokinase, alpha-D-glucopyranose, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
4EHZ
 
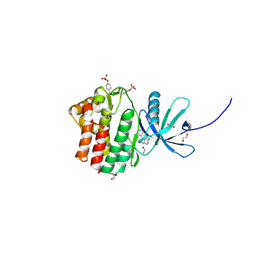 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-1-(piperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
