1H5U
 
 | | THE 1.76 A RESOLUTION CRYSTAL STRUCTURE OF GLYCOGEN PHOSPHORYLASE B COMPLEXED WITH GLUCOSE AND CP320626, A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Archontis, G. | | Deposit date: | 2001-05-25 | | Release date: | 2001-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The 1.76 A Resolution Crystal Structure of Glycogen Phosphorylase B Complexed with Glucose, and Cp320626, a Potential Antidiabetic Drug
Bioorg.Med.Chem., 10, 2002
|
|
4JL8
 
 | |
4JLA
 
 | |
1ISO
 
 | |
7XEY
 
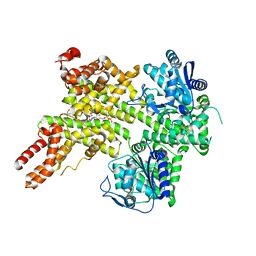 | | EDS1-PAD4 complexed with pRib-ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Huang, S, Jia, A, Xiao, Y. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
3DKV
 
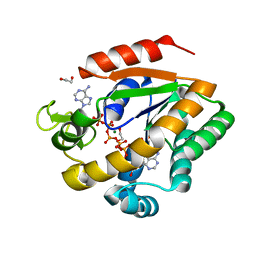 | | Crystal structure of adenylate kinase variant AKlse1 | | Descriptor: | 1,2-ETHANEDIOL, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, Bitto, E.B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of adenylate kinase variant AKlse1.
To be Published
|
|
4JKY
 
 | |
4JLB
 
 | |
3DL0
 
 | | Crystal structure of adenylate kinase variant AKlse3 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, McCoy, J.G. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
3CM0
 
 | | Crystal structure of adenylate kinase from Thermus thermophilus HB8 | | Descriptor: | Adenylate kinase | | Authors: | Nakagawa, N, Kondo, N, Masui, R, Matsuda, Z, Iwamoto, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-23 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenylate kinase from Thermus thermophilus HB8.
To be Published
|
|
3COV
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.5 Ang resolution- apo form | | Descriptor: | ETHANOL, GLYCEROL, Pantothenate synthetase, ... | | Authors: | Ciulli, A, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2008-03-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis pantothenate synthetase by analogues of the reaction intermediate.
Chembiochem, 9, 2008
|
|
8SUE
 
 | | Human asparagine synthetase (apo-ASNS) | | Descriptor: | Asparagine synthetase [glutamine-hydrolyzing] | | Authors: | Coricello, A, Zhu, W, Lupia, A, Gratteri, C, Vos, M, Chaptal, V, Alcaro, S, Takagi, Y, Richards, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Human asparagine synthetase (apo-ASNS)
To Be Published
|
|
2GN2
 
 | |
8ATD
 
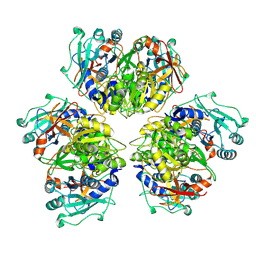 | | Wild type hexamer oxalyl-CoA synthetase (OCS) | | Descriptor: | Oxalate--CoA ligase | | Authors: | Lill, P, Burgi, J, Raunser, S, Wilmanns, M, Gatsogiannis, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol.Chem., 404, 2023
|
|
2GN1
 
 | |
2GN0
 
 | |
2P3S
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (G214R/Q199R) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Myers, J, Wu, G, Shamoo, Y. | | Deposit date: | 2007-03-09 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (G214R/Q199R)
To be Published
|
|
2ORI
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Shamoo, Y, Wilson, C.J, Wu, G, Myers, J. | | Deposit date: | 2007-02-02 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/)
To be Published
|
|
2Q2U
 
 | | Structure of Chlorella virus DNA ligase-product DNA complex | | Descriptor: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', Chlorella virus DNA ligase | | Authors: | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3OCZ
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase Complexed with the inhibitor adenosine 5-O-thiomonophosphate | | Descriptor: | ADENOSINE -5'-THIO-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of the inhibition of class C acid phosphatases by adenosine 5'-phosphorothioate.
Febs J., 278, 2011
|
|
2OO7
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Wu, G, Myers, J.C, Wittung-Stafshede, P, Shamoo, Y. | | Deposit date: | 2007-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R)
To be Published
|
|
2OSB
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Myers, J, Counago, R, Wilson, C.J, Wu, G, Shamoo, Y. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/)
To be Published
|
|
2QAJ
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q199R/G213E) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Myers, J, Wu, G, Shamoo, Y. | | Deposit date: | 2007-06-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q199R/G213E)
To be Published
|
|
1WXG
 
 | | E.coli NAD Synthetase, DND | | Descriptor: | MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXH
 
 | | E.coli NAD Synthetase, NAD | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
