8AWD
 
 | | Xylose Isomerase in 95% relative humidity environment | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8B3Q
 
 | |
8AWF
 
 | | Xylose Isomerase in 80% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWU
 
 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Xylose Isomerase with Glucose at 250ms | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWY
 
 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Serial measurement Xylose Isomerase with 2,3-butanediol at 50ms | | Descriptor: | MAGNESIUM ION, Meso-2,3-Butanediol, Xylose isomerase | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8B3P
 
 | |
8B3O
 
 | |
8AWB
 
 | | Xylose Isomerase in 90% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AJL
 
 | | Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
8AJA
 
 | | Structure of the Ancestral Scaffold Antigen-5 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
7XQH
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN-TM1i conformation) | | Descriptor: | C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQJ
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQI
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQB
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
8AFY
 
 | | Subtomogram average of membrane-bound Atg18 oligomers | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
7WVZ
 
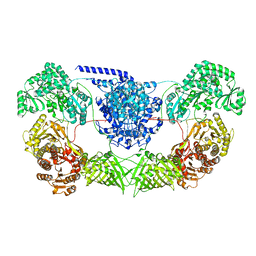 | | CalA3_modular PKS_KS-AT-DH-KR | | Descriptor: | Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
7X5A
 
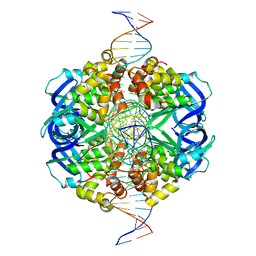 | | CryoEM structure of RuvA-Holliday junction complex | | Descriptor: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
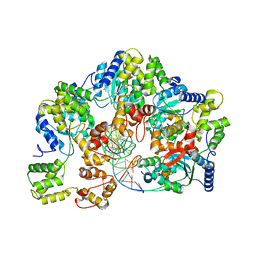 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
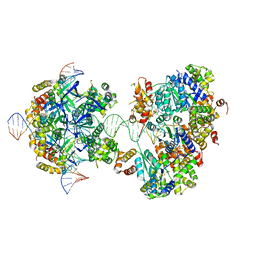 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
8B8A
 
 | | Multimerization domain of borna disease virus 1 phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bourhis, J.M, Chenavier, F, Freslon, L, Kawasaki, J, Horie, M, Tomonaga, K, Bachiri, K, Coyaud, E, Gonzalez-Dunia, D, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
7XKI
 
 | | Human Cx36/GJD2 (N-terminal deletion BRIL-fused mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein,Soluble cytochrome b562 | | Authors: | Cho, H.J, Lee, S.N, Jeong, H, Ryu, B, Lee, H.J, Woo, J.S, Lee, H.H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
6Q79
 
 | | Structure of Fucosylated D-antimicrobial peptide SB4 in complex with the Fucose-binding lectin PA-IIL at 2.009 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
7X6C
 
 | | Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class1 | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2022-03-07 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
8BTB
 
 | | Hexameric human IgG3 Fc complex | | Descriptor: | FLJ00385 protein (Fragment) | | Authors: | Abendstein, L, Sharp, T.H. | | Deposit date: | 2022-11-28 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Complement is activated by elevated IgG3 hexameric platforms and deposits C4b onto distinct antibody domains.
Nat Commun, 14, 2023
|
|
8BMQ
 
 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to AMP-PNP | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
