1IAQ
 
 | | C-H-RAS P21 PROTEIN MUTANT WITH THR 35 REPLACED BY SER (T35S) COMPLEXED WITH GUANOSINE-5'-[B,G-IMIDO] TRIPHOSPHATE | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Spoerner, M, Herrmann, C, Vetter, I.R, Kalbitzer, H.R, Wittinghofer, A. | | Deposit date: | 2001-03-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic properties of the Ras switch I region and its importance for binding to effectors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4A5W
 
 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
4A5T
 
 | | STRUCTURAL BASIS FOR THE CONFORMATIONAL MODULATION | | Descriptor: | CHLORIDE ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, PLASMINOGEN | | Authors: | Xue, Y, Bodin, C, Olsson, K. | | Deposit date: | 2011-10-28 | | Release date: | 2012-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structure of the Native Plasminogen Reveals an Activation-Resistant Compact Conformation.
J. Thromb. Haemost., 10, 2012
|
|
4AQ3
 
 | | HUMAN BCL-2 WITH PHENYLACYLSULFONAMIDE INHIBITOR | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, N,N-dibutyl-4-chloranyl-1-[2-(3,4-dihydro-1H-isoquinolin-2-ylcarbonyl)-4-[(7-iodanylnaphthalen-2-yl)sulfonylcarbamoyl]phenyl]-5-methyl-pyrazole-3-carboxamide | | Authors: | Bertrand, J.A, Fasolini, M, Modugno, M. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a Phenylacylsulfonamide Series of Dual Bcl-2/Bcl-Xl Antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4A1U
 
 | | Crystal structure of alpha-beta-foldamer 2c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1, CHLORIDE ION, ... | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
3ZZY
 
 | |
4B3Q
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, P51 RT, PRIMER DNA, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B1T
 
 | | Structure of the factor Xa-like trypsin variant triple-Ala (TA) in complex with eglin C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Menzel, A, Neumann, P, Stubbs, M.T. | | Deposit date: | 2012-07-12 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Thermodynamic signatures in macromolecular interactions involving conformational flexibility.
Biol.Chem., 395, 2014
|
|
8BD2
 
 | | Calcium-bound Calmodulin variant G113R | | Descriptor: | CALCIUM ION, Calmodulin-3 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Iwai, H, Niemelae, M.A, Brohus, M, Overgaard, M.T. | | Deposit date: | 2022-10-18 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
8BVC
 
 | |
8B3S
 
 | | Structure of YjbA in complex with ClpC N-terminal Domain | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC / Negative regulator of tic competence clcC/mecB, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Evans, N.J, Isaacson, R.L, Camp, A.H, Collins, M.K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of YjbA in complex with ClpC N-terminal Domain
To Be Published
|
|
5E0L
 
 | | LC8 - Chica (415-424) Complex | | Descriptor: | Dynein light chain 1, cytoplasmic, Protein Chica peptide, ... | | Authors: | Clark, S.A, Barbar, E.B, Karplus, P.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex.
Biochemistry, 55, 2016
|
|
8COK
 
 | |
8C6P
 
 | | Fragment screening hit I bound to endothiapepsin | | Descriptor: | 4-[(2-azanyl-4-methyl-1,3-thiazol-5-yl)methyl]benzenecarbonitrile, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C70
 
 | | Pyrrolidine fragment 1 bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(5-bromanylpyridin-3-yl)oxymethyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6Q
 
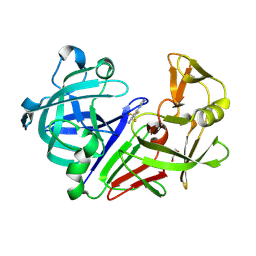 | | Fragment screening hit II bound to endothiapepsin | | Descriptor: | 3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)aniline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C71
 
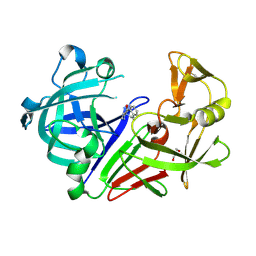 | | Pyrrolidine fragment 5b bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-(3,4-dihydro-1~{H}-isoquinolin-2-yl)pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C72
 
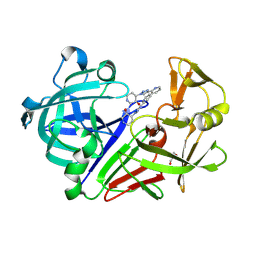 | | Pyrrolidine fragment 10b bound to endothiapepsin | | Descriptor: | (3~{S},4~{S})-4-(4-pyridin-2-yl-1,2,3-triazol-1-yl)piperidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C74
 
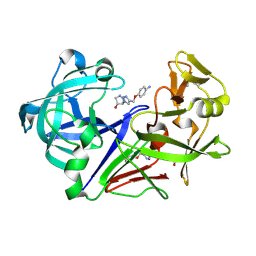 | | Pyrrolidine fragment 10d bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(4-azanylphenoxy)methyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6S
 
 | | Fragment screening hit III bound to endothiapepsin | | Descriptor: | 4-(1,4-diazepan-1-ylsulfonyl)isoquinoline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6T
 
 | | Fragment screening hit IV bound to endothiapepsin | | Descriptor: | 1-[3,5-bis(chloranyl)phenoxy]propan-2-amine, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
361D
 
 | | CRYSTAL STRUCTURE OF DOMAIN E OF THERMUS FLAVUS 5S RRNA: A HELICAL RNA-STRUCTURE INCLUDING A TETRALOOP | | Descriptor: | RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*GP*GP*CP*GP*AP*CP*CP*GP*CP*C P*UP*GP*G)-3') | | Authors: | Perbandt, M, Nolte, A, Lorenz, S, Erdmann, V.A, Betzel, C. | | Deposit date: | 1997-11-10 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of domain E of Thermus flavus 5S rRNA: a helical RNA structure including a hairpin loop.
FEBS Lett., 429, 1998
|
|
8CDG
 
 | | Crystal structure of human IL-17A cytokine in complex with macrocycle | | Descriptor: | (9~{S},12~{R},19~{S})-9-[[4-[[(2~{S})-2-cyclohexyl-2-(2-phenylethanoylamino)ethanoyl]amino]phenyl]methyl]-12-methyl-7,10,13,21-tetrakis(oxidanylidene)-8,11,14,20-tetrazaspiro[4.17]docosane-19-carboxylic acid, Interleukin-17A | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-01-30 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modulation of IL-17 backbone dynamics reduces receptor affinity and reveals a new inhibitory mechanism.
Chem Sci, 14, 2023
|
|
8BYV
 
 | | Cryo-EM structure of a Staphylococus aureus 30S-RbfA complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Bikmullin, A.G, Fatkhullin, B, Stetsenko, A, Guskov, A, Yusupov, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
8C3K
 
 | |
