8B1V
 
 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga | | Descriptor: | Dihydroprecondylocarpine acetate synthase 2, ZINC ION, precondylocarpine acetate | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1U9B
 
 | | MURINE/HUMAN UBIQUITIN-CONJUGATING ENZYME UBC9 | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E9 | | Authors: | Tong, H, Hateboer, G, Perrakis, A, Bernards, R, Sixma, T.K. | | Deposit date: | 1997-05-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system.
J.Biol.Chem., 272, 1997
|
|
1U2Q
 
 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Weight Protein Tyrosine Phosphatase (MPtpA) at 2.5A resolution with glycerol in the active site | | Descriptor: | CHLORIDE ION, GLYCEROL, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
8B0I
 
 | | CryoEM structure of bacterial RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small regulatory RNA, RNase adapter protein RapZ | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
3S7T
 
 | |
1U2V
 
 | | Crystal structure of Arp2/3 complex with bound ADP and calcium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-Related Protein 2, Actin-Related Protein 3, ... | | Authors: | Nolen, B.J, Littlefield, R.S, Pollard, T.D. | | Deposit date: | 2004-07-20 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8B11
 
 | | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A and CsoS2 co-expression (T = 4) | | Descriptor: | Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Ni, T, Jiang, Q, Liu, L.N, Zhang, P. | | Deposit date: | 2022-09-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Intrinsically disordered CsoS2 acts as a general molecular thread for alpha-carboxysome shell assembly.
Nat Commun, 14, 2023
|
|
3S4P
 
 | | Crystal structure of the bacterial ribosomal decoding site complexed with an amphiphilic paromomycin O2''-ether analogue | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-2-O-{2-[(2-phenylethyl)amino]ethyl}-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Szychowski, J, Kondo, J, Zahr, O, Auclair, K, Westhof, E, Hanessian, S, Keillor, J.W. | | Deposit date: | 2011-05-20 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Inhibition of aminoglycoside-deactivating enzymes APH(3')-IIIa and AAC(6')-Ii by amphiphilic paromomycin O2''-ether analogues
Chemmedchem, 6, 2011
|
|
1U3U
 
 | |
8AT3
 
 | | Structure of the augmin holocomplex in open conformation | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, HAUS augmin like complex subunit 4 L homeolog, HAUS augmin like complex subunit 6 L homeolog, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (33 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
8B55
 
 | | Human ADGRG4 PTX-like domain | | Descriptor: | Adhesion G-protein coupled receptor G4, MAGNESIUM ION | | Authors: | Kieslich, B, Straeter, N. | | Deposit date: | 2022-09-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J.Biol.Chem., 299, 2023
|
|
3S5R
 
 | |
2IPT
 
 | | PFA1 Fab Fragment | | Descriptor: | ACETAMIDE, IgG2a Fab fragment Heavy Chain, IgG2a Fab fragment Light Chain Kappa | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2006-10-12 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1U9O
 
 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
1U9Y
 
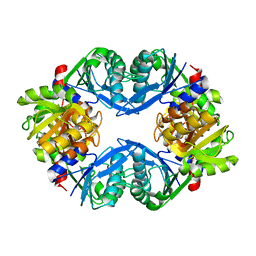 | | Crystal Structure of Phosphoribosyl Diphosphate Synthase from Methanocaldococcus jannaschii | | Descriptor: | Ribose-phosphate pyrophosphokinase | | Authors: | Kadziola, A, Johansson, E, Jepsen, C.H, McGuire, J, Larsen, S, Hove-Jensen, B. | | Deposit date: | 2004-08-11 | | Release date: | 2005-08-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii
J.Mol.Biol., 354, 2005
|
|
8B25
 
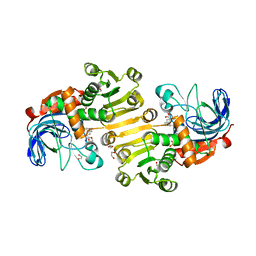 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga - stemmadenine acetate bound structure | | Descriptor: | 1,2-ETHANEDIOL, Dihydroprecondylocarpine acetate synthase 2, SULFATE ION, ... | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1UAQ
 
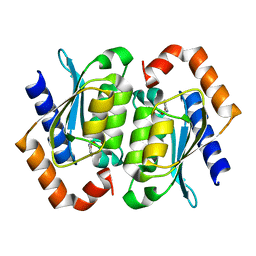 | | The crystal structure of yeast cytosine deaminase | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, ZINC ION, cytosine deaminase | | Authors: | Ko, T.-P, Lin, J.-J, Hu, C.-Y, Hsu, Y.-H, Wang, A.H.-J, Liaw, S.-H. | | Deposit date: | 2003-03-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution
J.Biol.Chem., 278, 2003
|
|
1U4F
 
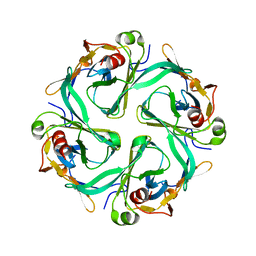 | | Crystal Structure of Cytoplasmic Domains of IRK1 (Kir2.1) channel | | Descriptor: | Inward rectifier potassium channel 2 | | Authors: | Pegan, S, Arrabit, C, Zhou, W, Kwiatkowski, W, Slesinger, P.A, Choe, S. | | Deposit date: | 2004-07-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification
Nat.Neurosci., 8, 2005
|
|
8B2R
 
 | |
8B81
 
 | | The structure of Gan1D W433A in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | The structure of Gan1D W433A in complex with cellobiose-6-phosphate
To Be Published
|
|
3S91
 
 | | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain-containing protein 3, ISOPROPYL ALCOHOL | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1
To be Published
|
|
1U5D
 
 | | Crystal Structure of the PH domain of SKAP55 | | Descriptor: | SULFATE ION, Src Kinase-associated Phosphoprotein of 55 kDa | | Authors: | Tang, Y, Swanson, K.D, Neel, B.G, Eck, M.J. | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Dimerization and Phosphoinositide Specificity of the Src Kinase-associated Phosphoproteins SKAP55 and SKAP-Hom
To be Published
|
|
1U5U
 
 | |
8B80
 
 | | The structure of Gan1D W433A in complex with galactose-6P | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of Gan1D W433A in complex with galactose-6P
To Be Published
|
|
1U6D
 
 | |
