2XC2
 
 | | Crystal structure of oxidized Schistosoma mansoni Thioredoxin pre- protein at 1.6 Angstrom | | Descriptor: | CALCIUM ION, THIOREDOXINN, ZINC ION | | Authors: | Boumis, G, Miele, A.E, Dimastrogiovanni, D, Angelucci, F, Bellelli, A. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Functional Characterization of Schistosoma Mansoni Thioredoxin.
Protein Sci., 20, 2011
|
|
2XNR
 
 | |
2XBQ
 
 | | Crystal structure of reduced Schistosoma mansoni Thioredoxin pre- protein at 1.7 Angstrom | | Descriptor: | THIOREDOXIN, ZINC ION | | Authors: | Boumis, G, Miele, A.E, Dimastrogiovanni, D, Angelucci, F, Bellelli, A. | | Deposit date: | 2010-04-14 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Functional Characterization of Schistosoma Mansoni Thioredoxin.
Protein Sci., 20, 2011
|
|
2Y5Z
 
 | | Mixed-function P450 MycG in complex with mycinamicin III in C2221 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN III, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2XNQ
 
 | |
2XPN
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form I | | Descriptor: | BROMIDE ION, CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2Y5N
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin V in P21 space group | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYCINAMICIN V, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y4X
 
 | |
2Y98
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in P21212 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-02-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2ZQ2
 
 | | Exploring trypsin S3 pocket | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-08-03 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
2XPP
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form III | | Descriptor: | CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2ZQ1
 
 | | Exploring trypsin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-08-03 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
2XTB
 
 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Activator | | Descriptor: | 4-[5-(4-PHENOXYPHENYL)-1H-PYRAZOL-3-YL]MORPHOLINE, ADENOSINE KINASE | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of T. B. Rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation.
Plos Negl Trop Dis, 5, 2011
|
|
2XP1
 
 | | Structure of the tandem SH2 domains from Antonospora locustae transcription elongation factor Spt6 | | Descriptor: | CHLORIDE ION, SPT6, SULFATE ION | | Authors: | Diebold, M.-L, Koch, M, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Noncanonical Tandem Sh2 Enables Interaction of Elongation Factor Spt6 with RNA Polymerase II.
J.Biol.Chem., 285, 2010
|
|
2YCA
 
 | | Mixed-function P450 MycG in complex with mycinamicin III in P21212 space group | | Descriptor: | GLYCEROL, MYCINAMICIN III, P-450-LIKE PROTEIN, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
1TLJ
 
 | | Crystal Structure of Conserved Protein of Unknown Function SSO0622 from Sulfolobus solfataricus | | Descriptor: | Hypothetical UPF0130 protein SSO0622, SULFATE ION | | Authors: | Jia, Z, Wong, A.H.Y, Kudrytska, M, Skarina, T, Walker, J, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of the TYW3/Taw3 class of SAM-dependent methyltransferases.
Rna, 23, 2017
|
|
1P0S
 
 | | Crystal Structure of Blood Coagulation Factor Xa in Complex with Ecotin M84R | | Descriptor: | Coagulation factor X precursor, Ecotin precursor, MAGNESIUM ION, ... | | Authors: | Wang, S.X, Hur, E, Sousa, C.A, Brinen, L, Slivka, E.J, Fletterick, R.J. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Extended Interactions and Gla Domain of Blood Coagulation Factor Xa
Biochemistry, 42, 2003
|
|
2ZHD
 
 | | Exploring trypsin S3 pocket | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
1TI1
 
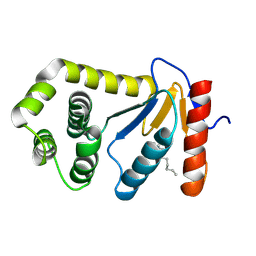 | | crystal structure of a mutant DsbA | | Descriptor: | DODECANE, Thiol:disulfide interchange protein dsbA | | Authors: | Kone, A, Serre, L. | | Deposit date: | 2004-06-02 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intriguing conformation changes associated with the trans/cis isomerization of a prolyl residue in the active site of the DsbA C33A mutant.
J.Mol.Biol., 347, 2005
|
|
1PIN
 
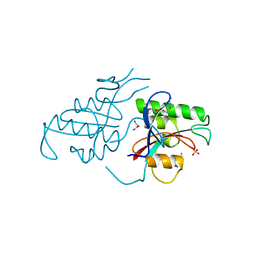 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Noel, J.P, Ranganathan, R, Hunter, T. | | Deposit date: | 1998-06-21 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
1TMR
 
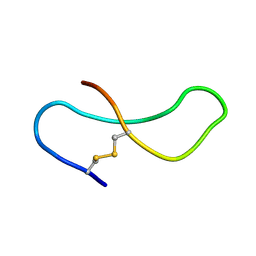 | | THE STRUCTURE OF A 19 RESIDUE FRAGMENT FROM THE C-LOOP OF THE FOURTH EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF THROMBOMODULIN | | Descriptor: | THROMBOMODULIN PRECURSOR | | Authors: | Adler, M, Seto, M, Nitecki, D, Light, D.R, Morser, J. | | Deposit date: | 1994-05-20 | | Release date: | 1995-06-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of a 19-residue fragment from the C-loop of the fourth epidermal growth factor-like domain of thrombomodulin.
J.Biol.Chem., 270, 1995
|
|
1U5F
 
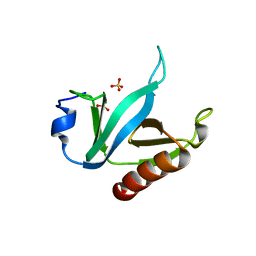 | |
1Z81
 
 | | Crystal Structure of cyclophilin from Plasmodium yoelii. | | Descriptor: | cyclophilin | | Authors: | Mulichak, A, Alam, Z, Amani, M, Lew, J, Wasney, G, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-29 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1ZML
 
 | | Crystal Structure of the Catalytic Domain of Factor XI in complex with (R)-1-(4-(4-(hydroxymethyl)-1,3,2-dioxaborolan-2-yl)phenethyl)guanidine | | Descriptor: | (R)-1-(4-(4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL)PHENETHYL)GUANIDINE, BICARBONATE ION, Coagulation factor XI, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M.J, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R.E, Strickler, J.E. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2A7Y
 
 | | Solution Structure of the Conserved Hypothetical Protein Rv2302 from the Bacterium Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv2302/MT2359 | | Authors: | Buchko, G.W, Kim, C.-Y, Terwilliger, T.C, Kennedy, M.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-06 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein Rv2302 from Mycobacterium tuberculosis.
J.Bacteriol., 188, 2006
|
|
