8VCI
 
 | | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop | | Descriptor: | Frameshift Stimulatory Element with Upstream Multi-branch Loop | | Authors: | Peterson, J.M, Becker, S.T, O'Leary, C.A, Juneja, P, Yang, Y, Moss, W.N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop.
Biochemistry, 63, 2024
|
|
4MKF
 
 | | Crystal structure of a stable adenylate kinase variant AKv3 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CALCIUM ION, ... | | Authors: | Moon, S, Jung, D, Bae, E. | | Deposit date: | 2013-09-05 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An integrated approach for thermal stabilization of a mesophilic adenylate kinase.
Proteins, 82, 2014
|
|
4MKG
 
 | | Crystal structure of a stable adenylate kinase variant AKv8 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moon, S, Jung, D, Bae, E. | | Deposit date: | 2013-09-05 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An integrated approach for thermal stabilization of a mesophilic adenylate kinase.
Proteins, 82, 2014
|
|
4NDI
 
 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDH
 
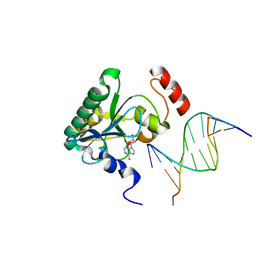 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDF
 
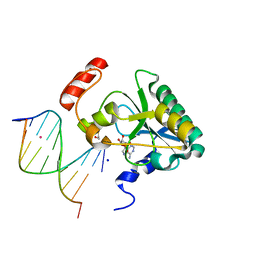 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
8C10
 
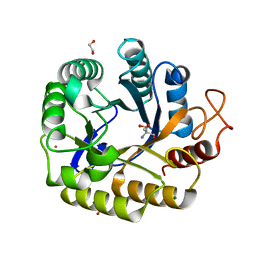 | | Biochemical and structural characterisation of an alkaline family GH5 cellulase from a shipworm symbiont | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 Cellulase, ... | | Authors: | Leiros, I, Vaaje-Kolstad, G. | | Deposit date: | 2022-12-19 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Biochemical and structural characterisation of a family GH5 cellulase from endosymbiont of shipworm P. megotara.
Biotechnol Biofuels Bioprod, 16, 2023
|
|
6DTL
 
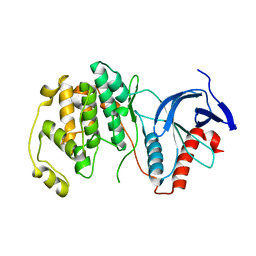 | | Mitogen-activated protein kinase 6 | | Descriptor: | Mitogen-activated protein kinase 6 | | Authors: | Ruble, J. | | Deposit date: | 2018-06-17 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to SPEECHLESS.
Nat.Plants, 5, 2019
|
|
4NDG
 
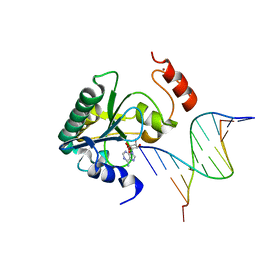 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
2WY2
 
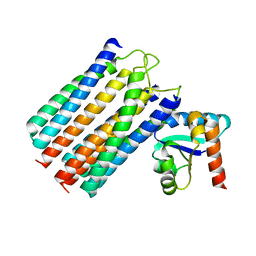 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2WWV
 
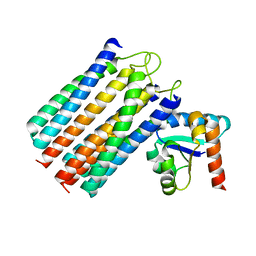 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
6ZL2
 
 | |
6ZL9
 
 | |
6ZTE
 
 | |
3I1D
 
 | | Distinct recognition of three-way DNA junctions by the two enantiomers of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Boer, D.R, Uson, I, Hannon, M.J, Coll, M. | | Deposit date: | 2009-06-26 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
4HN7
 
 | |
7NKW
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
4J3R
 
 | | Crystal structure of catechol oxidase from Aspergillus oryzae, soaked in 4-tert-butylcatechol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Hakulinen, N, Gasparetti, C, Kaljunen, H, Rouvinen, J. | | Deposit date: | 2013-02-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of an extracellular catechol oxidase from the ascomycete fungus Aspergillus oryzae.
J.Biol.Inorg.Chem., 18, 2013
|
|
4IQZ
 
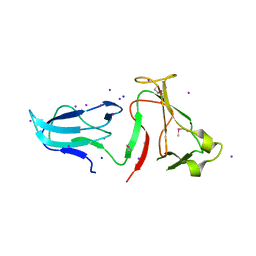 | | The crystal structure of a large insert in RNA polymerase (RpoC) subunit from E. coli | | Descriptor: | DNA-directed RNA polymerase subunit beta', IODIDE ION, SODIUM ION | | Authors: | Bhandari, V, Sugiman-Marangos, S.N, Naushad, H.S, Gupta, R.S, Junop, M.S. | | Deposit date: | 2013-01-14 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a large insert in RNA polymerase (RpoC) subunit from E. coli
To be Published
|
|
4R9S
 
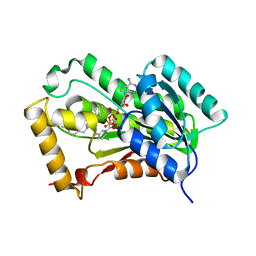 | | Mycobacterium tuberculosis InhA bound to NITD-916 | | Descriptor: | 6-[(4,4-dimethylcyclohexyl)methyl]-4-hydroxy-3-phenylpyridin-2(1H)-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Noble, C.G. | | Deposit date: | 2014-09-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Direct inhibitors of InhA are active against Mycobacterium tuberculosis
Sci Transl Med, 7, 2015
|
|
4IPV
 
 | |
4IPU
 
 | |
4R9R
 
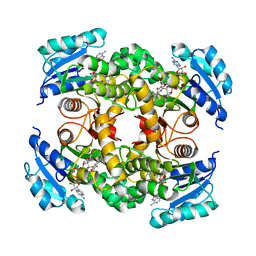 | | Mycobacterium tuberculosis InhA bound to NITD-564 | | Descriptor: | 6-(cyclohexylmethyl)-4-hydroxy-3-phenylpyridin-2(1H)-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Noble, C.G. | | Deposit date: | 2014-09-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Direct inhibitors of InhA are active against Mycobacterium tuberculosis
Sci Transl Med, 7, 2015
|
|
1JDY
 
 | |
4JDR
 
 | | Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase from escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chandrasekhar, K, Arjunan, P, Furey, W. | | Deposit date: | 2013-02-25 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight to the Interaction of the Dihydrolipoamide Acetyltransferase (E2) Core with the Peripheral Components in the Escherichia coli Pyruvate Dehydrogenase Complex via Multifaceted Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
