3KE2
 
 | |
3KE7
 
 | |
3KD4
 
 | |
3KEN
 
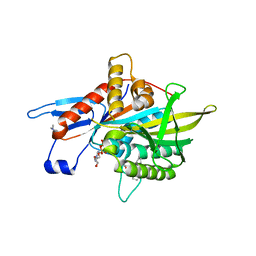 | | Human Eg5 in complex with S-trityl-L-cysteine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Parke, C.L, Wojcik, E.J, Kim, S, Worthylake, D.K. | | Deposit date: | 2009-10-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core.
J.Biol.Chem., 285, 2010
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3MSW
 
 | |
3MBH
 
 | |
3M81
 
 | |
3MDQ
 
 | |
3MX8
 
 | | Crystal structure of ribonuclease A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, LINKER, ... | | Authors: | Leich, F, Neumann, P, Lilie, H, Ulbrich-Hofmann, R, Arnold, U. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RNase A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor
Febs J., 278, 2011
|
|
3N0W
 
 | |
3MWQ
 
 | |
3MX3
 
 | |
3MTQ
 
 | |
3MZ2
 
 | |
3N0X
 
 | |
3N0Q
 
 | |
3MHM
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[N-(6-benzylamino-5-nitropyrimidin-4-yl)amino]methyl}benzenesulfonamide | | Descriptor: | 4-({[6-(benzylamino)-5-nitropyrimidin-4-yl]amino}methyl)benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-04-08 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3M96
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 5-{[(5-bromo-1H-benzimidazol-2-yl)sulfanyl]acetyl}-2-chlorobenzenesulfonamide | | Descriptor: | 5-{[(5-bromo-1H-benzimidazol-2-yl)sulfanyl]acetyl}-2-chlorobenzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-20 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII.
Bioorg.Med.Chem., 18, 2010
|
|
3MEI
 
 | | Regulatory motif from the thymidylate synthase mRNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*GP*CP*GP*CP*CP*AP*(5BU)P*GP*CP*CP*UP*GP*UP*GP*GP*CP*GP*G)-3') | | Authors: | Dibrov, S, McLean, J, Hermann, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Structure of an RNA dimer of a regulatory element from human thymidylate synthase mRNA.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3MG1
 
 | | Crystal structure of the orange carotenoid protein from cyanobacteria Synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3MSQ
 
 | |
3M1V
 
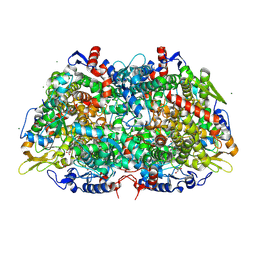 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-05 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
3M67
 
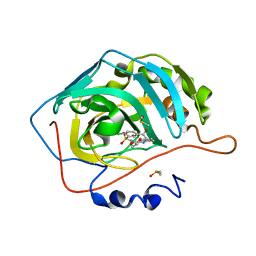 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3MG3
 
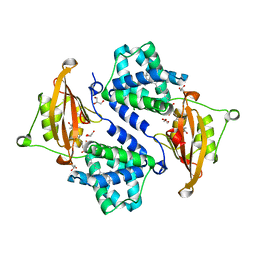 | | Crystal structure of the orange carotenoid protein R155L mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
