1HW5
 
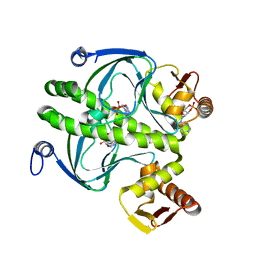 | | THE CAP/CRP VARIANT T127L/S128A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | Authors: | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
4ENN
 
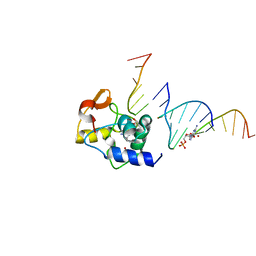 | |
7V97
 
 | | Arsenic-bound p53 DNA-binding domain mutant V272M | | Descriptor: | ARSENIC, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Xing, Y.F, Wang, Z.Y, Ni, Y, Song, H.X. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Diverse rescue potencies of p53 mutations to ATO are predetermined by intrinsic mutational properties.
Sci Transl Med, 15, 2023
|
|
5YIU
 
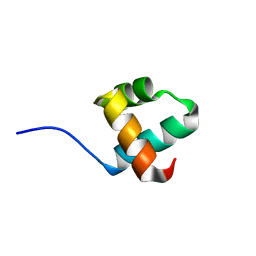 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) | | Descriptor: | Cell cycle regulatory protein GcrA | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
6BC8
 
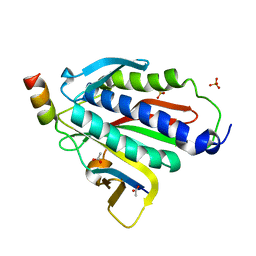 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | Descriptor: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1BW6
 
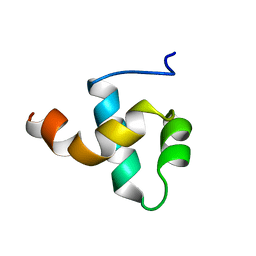 | | HUMAN CENTROMERE PROTEIN B (CENP-B) DNA BINDIGN DOMAIN RP1 | | Descriptor: | PROTEIN (CENTROMERE PROTEIN B) | | Authors: | Iwahara, J, Kigawa, T, Kitagawa, K, Masumoto, H, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-09-30 | | Release date: | 1998-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A helix-turn-helix structure unit in human centromere protein B (CENP-B).
EMBO J., 17, 1998
|
|
4U63
 
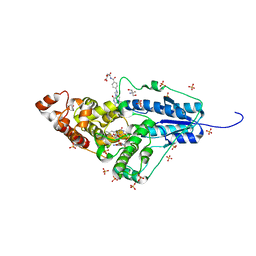 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
6ZWT
 
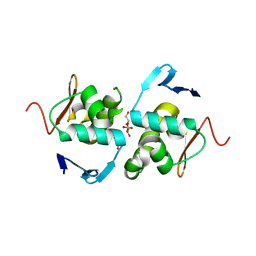 | | Crystal structure of DNA-binding domain of OmpR of two-component system of Acinetobacter baumannii | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Lucchini, V, Trebosc, V, Gartenmann, S, Pieren, M, Kemmer, C, Kammerer, R.A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis.
Virulence, 13, 2022
|
|
1QLN
 
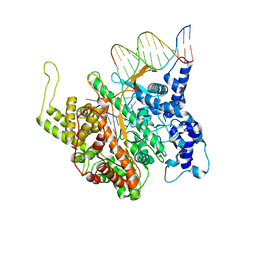 | | STRUCTURE OF A TRANSCRIBING T7 RNA POLYMERASE INITIATION COMPLEX | | Descriptor: | BACTERIOPHAGE T7 RNA POLYMERASE, DNA (5- D (P*CP*TP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP* AP*TP*TP*A)-3), DNA (5-D(P*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*TP*A)-3), ... | | Authors: | Cheetham, G.M.T, Steitz, T.A. | | Deposit date: | 1999-09-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Transcribing T7 RNA Polymerase Initiation Complex
Science, 286, 1999
|
|
1S0V
 
 | | Structural basis for substrate selection by T7 RNA polymerase | | Descriptor: | 5'-D(*G*GP*GP*AP*AP*TP*CP*GP*AP*TP*AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*C)-3', 5'-R(*AP*AP*CP*U*GP*CP*GP*GP*CP*GP*AP*U)-3', ... | | Authors: | Temiakov, D, Patlan, V, Anikin, M, McAllister, W.T, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate selection by t7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
4IPC
 
 | | Structure of the N-terminal domain of RPA70, E7R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPD
 
 | | Structure of the N-terminal domain of RPA70, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPG
 
 | | Structure of the N-terminal domain of RPA70, E7R, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPH
 
 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
3U3W
 
 | | Crystal Structure of Bacillus thuringiensis PlcR in complex with the peptide PapR7 and DNA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*AP*AP*AP*TP*AP*TP*TP*GP*CP*AP*TP*AP*G)-3', 5'-D(P*CP*TP*AP*TP*GP*CP*AP*AP*TP*AP*TP*TP*TP*CP*AP*TP*AP*T)-3', C-terminus heptapeptide from PapR protein, ... | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2011-10-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1AE3
 
 | | MUTANT R82C OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
7K78
 
 | | antibody and nucleosome complex | | Descriptor: | Cse4, DNA (136-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
1AE2
 
 | | MUTANT L32R OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
6MKM
 
 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MK3
 
 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MKK
 
 | | Crystallographic solvent mapping analysis of DMSO/Mg bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
5HDN
 
 | |
6ZJ2
 
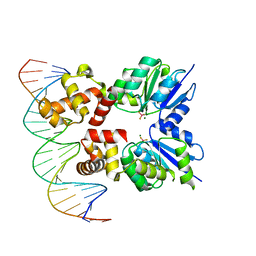 | | Structure of RcsB from Salmonella enterica serovar Typhimurium bound to promoter rprA in the presence of phosphomimetic BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(P*CP*CP*GP*AP*TP*CP*AP*GP*AP*TP*TP*CP*GP*TP*CP*TP*CP*AP*AP*TP*AP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Huesa, J, Marina, A, Casino, P. | | Deposit date: | 2020-06-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon.
Nucleic Acids Res., 49, 2021
|
|
2O5I
 
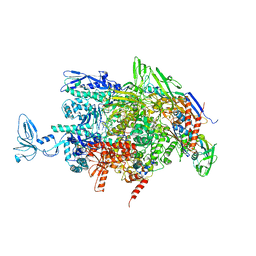 | | Crystal structure of the T. thermophilus RNA polymerase elongation complex | | Descriptor: | 5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3', 5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*GP*AP*GP*UP*CP*UP*GP*CP*GP*GP*CP*GP*CP*GP*CP*G)-3', ... | | Authors: | Vassylyev, D.G, Tahirov, T.H, Vassylyeva, M.N. | | Deposit date: | 2006-12-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for transcription elongation by bacterial RNA polymerase.
Nature, 448, 2007
|
|
