7NYT
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with lactose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-03-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
6AP7
 
 | |
7OFK
 
 | | Ligand complex of RORg LBD | | Descriptor: | (1~{R})-2-ethanoyl-~{N}-[4-[1,1,1,3,3,3-hexakis(fluoranyl)-2-oxidanyl-propan-2-yl]phenyl]-5-methylsulfonyl-1,3-dihydroisoindole-1-carboxamide, DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | AZD0284, a Potent, Selective, and Orally Bioavailable Inverse Agonist of Retinoic Acid Receptor-Related Orphan Receptor C2.
J.Med.Chem., 64, 2021
|
|
5BXB
 
 | |
7WNN
 
 | |
7SMH
 
 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
6PII
 
 | | The evolving story of AtzT, a periplasmic binding protein | | Descriptor: | Atrazine periplasmic binding protein, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6APA
 
 | |
6APW
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 4-[(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)methyl]benzoic acid, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
8D1C
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-(Trifluoromethoxy)benzoic acid | | Descriptor: | 4-(trifluoromethoxy)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-05-27 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Oxidations Using a Cytochrome P450 Enzyme Variant Driven with Surrogate Oxygen Donors and Light.
Chemistry, 28, 2022
|
|
7SNW
 
 | | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576 | | Descriptor: | 2-(methoxycarbonyl)thiophene-3-sulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576
To be published
|
|
7NUE
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
6AQK
 
 | |
7OFI
 
 | | Ligand complex of RORg LBD | | Descriptor: | (2~{R})-2-(4-ethylsulfonylphenyl)-~{N}-[4-[1,1,1,3,3,3-hexakis(fluoranyl)-2-oxidanyl-propan-2-yl]phenyl]-~{N}'-methyl-butanediamide, DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | AZD0284, a Potent, Selective, and Orally Bioavailable Inverse Agonist of Retinoic Acid Receptor-Related Orphan Receptor C2.
J.Med.Chem., 64, 2021
|
|
6P6Q
 
 | | HCV NS3/4A protease domain of genotype 1a3a chimera in complex with grazoprevir | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 4A,Serine protease NS3, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
6AR1
 
 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Nat)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
6ARJ
 
 | | Crystal structure of CARM1 with EPZ022302 and SAH | | Descriptor: | GLYCEROL, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.
Sci Rep, 7, 2017
|
|
6PJA
 
 | |
6AQR
 
 | | SAGA DUB module Ubp8(C146A)/Sgf11/Sus1/Sgf73 bound to monoubiquitin | | Descriptor: | Polyubiquitin-C, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Morrow, M.E, Morgan, M.T, Wolberger, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins.
EMBO Rep., 19, 2018
|
|
7NR6
 
 | | Structure of NUDT15 in complex with NSC56456 | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-9-cyclohexyl-3~{H}-purine-6-thione, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coupling cellular target engagement to drug-induced responses with CeTEAM
To Be Published
|
|
8Q5E
 
 | |
8DF1
 
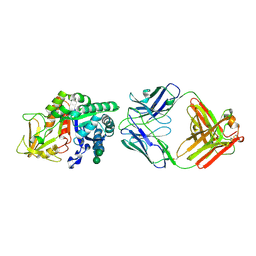 | | Chi3l1 bound by antibody C59 | | Descriptor: | C59 Fab Heavy chain, C59 Fab Light chain, Chitinase-3-like protein 1, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A novel humanized Chi3l1 blocking antibody attenuates acetaminophen-induced liver injury in mice.
Antib Ther, 6, 2023
|
|
6V7J
 
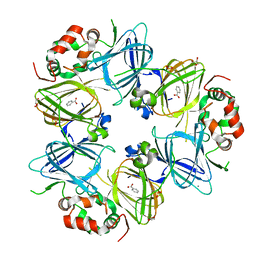 | | The C2221 crystal form of canavalin at 173 K | | Descriptor: | BENZOIC ACID, CALCIUM ION, Canavalin, ... | | Authors: | McPherson, A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of benzoic acid and anions within the cupin domains of the vicilin protein canavalin from jack bean (Canavalia ensiformis): Crystal structures.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6MAC
 
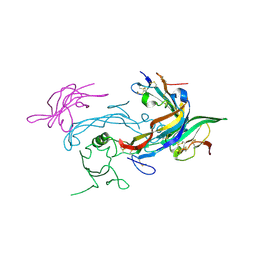 | | Ternary structure of GDF11 bound to ActRIIB-ECD and Alk5-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 11, ... | | Authors: | Goebel, E.J, Thompson, T.B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7SQI
 
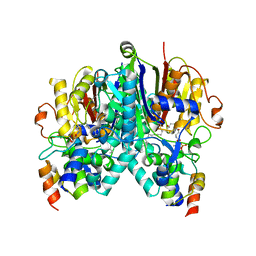 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-05 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
