2BBN
 
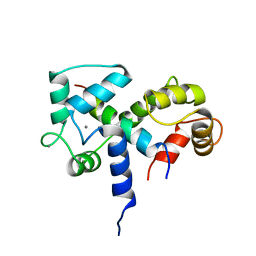 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
1HEB
 
 | |
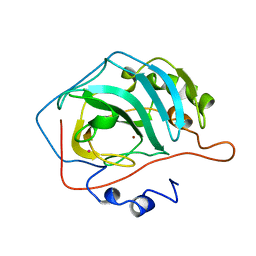
1HEA
 
 | |
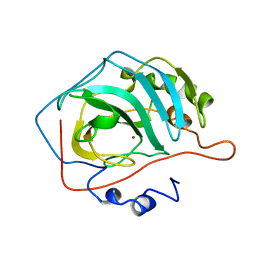
1HEC
 
 | |
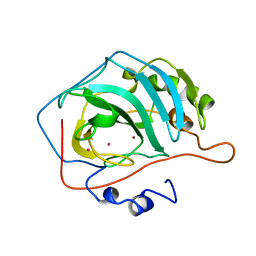
1HED
 
 | |
2SNV
 
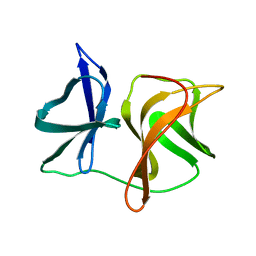 | |
1D89
 
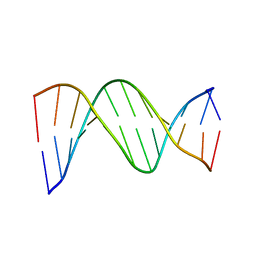 | |
2AT2
 
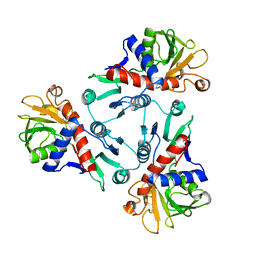 | |
1TLK
 
 | |
1MDC
 
 | |
1GRC
 
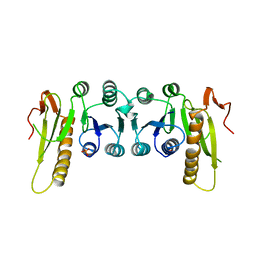 | |
1D83
 
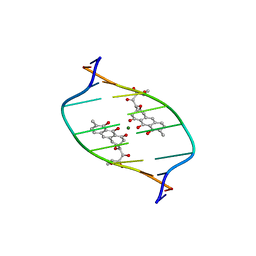 | | STRUCTURE REFINEMENT OF THE CHROMOMYCIN DIMER/DNA OLIGOMER COMPLEX IN SOLUTION | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Gao, X, Mirau, P, Patel, D.J. | | Deposit date: | 1992-07-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the chromomycin dimer-DNA oligomer complex in solution.
J.Mol.Biol., 223, 1992
|
|
1PTS
 
 | |
1PHD
 
 | |
1PHG
 
 | |
1PHC
 
 | |
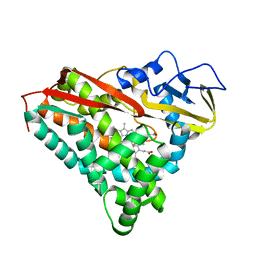
1PHF
 
 | |
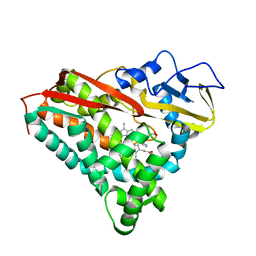
1PHE
 
 | |
1PHB
 
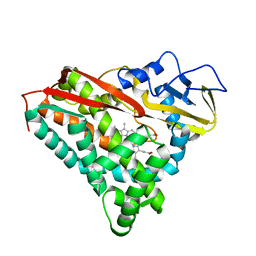
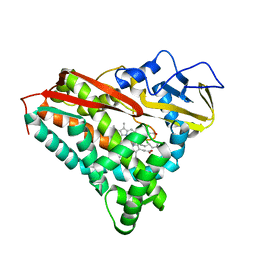 | | INHIBITOR-INDUCED CONFORMATIONAL CHANGE IN CYTOCHROME P450-CAM | | Descriptor: | 1-(N-IMIDAZOLYL)-2-HYDROXY-2-(2,3-DICHLOROPHENYL)OCTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Poulos, T.L. | | Deposit date: | 1992-07-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitor-induced conformational change in cytochrome P-450CAM.
Biochemistry, 32, 1993
|
|
1PHA
 
 | | INHIBITOR-INDUCED CONFORMATIONAL CHANGE IN CYTOCHROME P450-CAM | | Descriptor: | 1-(N-IMIDAZOLYL)-2-HYDROXY-2-(2,3-DICHLOROPHENYL)OCTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Poulos, T.L. | | Deposit date: | 1992-07-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Inhibitor-induced conformational change in cytochrome P-450CAM.
Biochemistry, 32, 1993
|
|
1BRP
 
 | |
1BRQ
 
 | |
6Q21
 
 | |
1THG
 
 | | 1.8 ANGSTROMS REFINED STRUCTURE OF THE LIPASE FROM GEOTRICHUM CANDIDUM | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrag, J.D, Cygler, M. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A refined structure of the lipase from Geotrichum candidum.
J.Mol.Biol., 230, 1993
|
|
1MRR
 
 | |
