6JVH
 
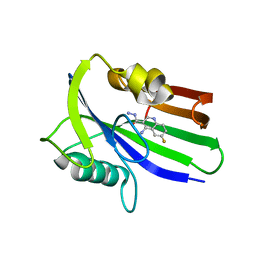 | | Crystal structure of human MTH1 in complex with compound MI0320 | | Descriptor: | 4-amino-6-fluoroquinoline-3-carbohydrazide, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
5SCV
 
 | |
6JVO
 
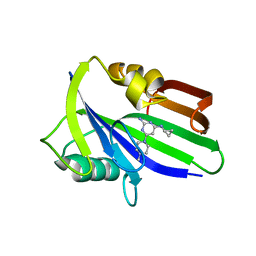 | | Crystal structure of human MTH1 in complex with compound MI1022 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
5SD5
 
 | |
6KEK
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6-hydroxy-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | Descriptor: | 6-hydroxy-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KER
 
 | | Crystal structure of D113A mutant of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in glutathione-bound form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KF7
 
 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 mutant S285G
To Be Published
|
|
6KFS
 
 | | Structure of CCM related protein | | Descriptor: | LCIB_C_CA domain-containing protein, ZINC ION | | Authors: | Jin, S, Gao, Y.G. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biochemical characterization of novel carbonic anhydrases from Phaeodactylum tricornutum.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5SDA
 
 | |
5SD7
 
 | |
6JVY
 
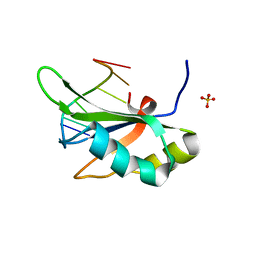 | | Crystal structure of RBM38 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*GP*TP*GP*TP*GP*TP*GP*TP*G)-3'), RNA-binding protein 38, SULFATE ION | | Authors: | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
6JW6
 
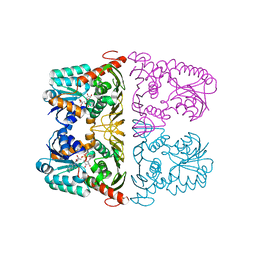 | | The crystal structure of KanD2 in complex with NAD | | Descriptor: | Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
5SD1
 
 | |
6JWM
 
 | |
5SCX
 
 | |
6KFV
 
 | |
5SD9
 
 | |
5SCO
 
 | |
6KI2
 
 | |
6KI8
 
 | | Pyrophosphatase mutant K149R from Acinetobacter baumannii | | Descriptor: | DIPHOSPHATE, Inorganic pyrophosphatase, MAGNESIUM ION | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
5SCM
 
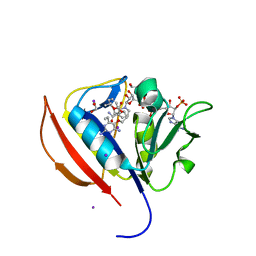 | |
6KIJ
 
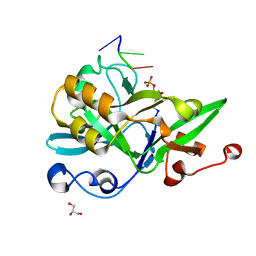 | | Crystal structure of yedK with ssDNA containing an abasic site | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*CP*GP*TP*CP*G)-3'), GLYCEROL, PENTANE-3,4-DIOL-5-PHOSPHATE, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-07-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
5SDB
 
 | |
6JXJ
 
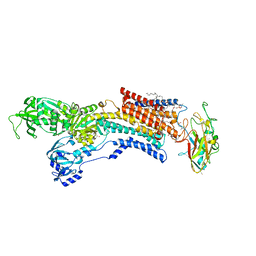 | | Rb+-bound E2-AlF state of the gastric proton pump (Tyr799Trp) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Irie, K. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A single K + -binding site in the crystal structure of the gastric proton pump.
Elife, 8, 2019
|
|
5SCP
 
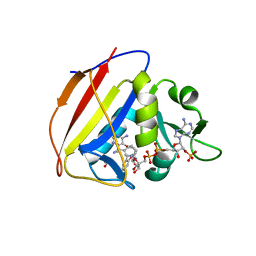 | |
