5HYG
 
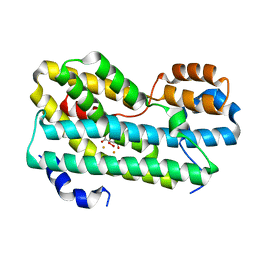 | | CmlI (peroxo bound state), arylamine oxygenase of chloramphenicol biosynthetic pathway | | Descriptor: | FE (III) ION, HYDROGEN PEROXIDE, L(+)-TARTARIC ACID, ... | | Authors: | Knoot, C.J, Lipscomb, J.D. | | Deposit date: | 2016-02-01 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of CmlI, the arylamine oxygenase from the chloramphenicol biosynthetic pathway.
J.Biol.Inorg.Chem., 21, 2016
|
|
5HYH
 
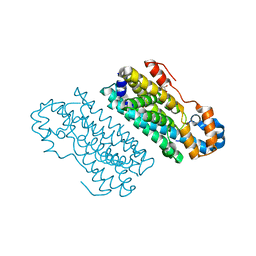 | |
7V06
 
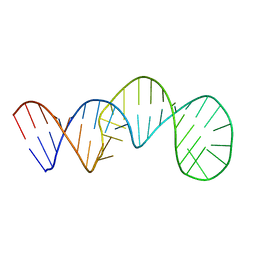 | |
6BM9
 
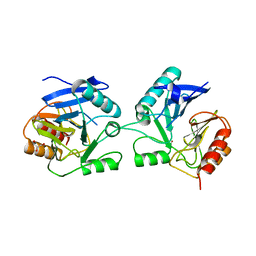 | |
9J7T
 
 | |
9IZ6
 
 | |
6E1Y
 
 | |
8ZI2
 
 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
1J41
 
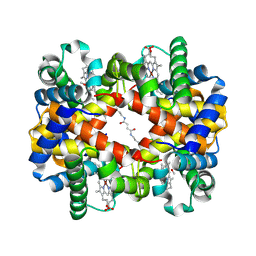 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Ni)-beta(Fe) hemoglobin (laser photolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8ZI0
 
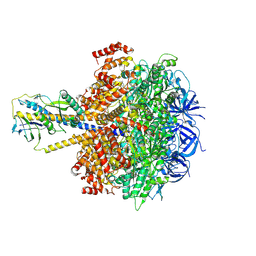 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
8ZI1
 
 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
8ZI3
 
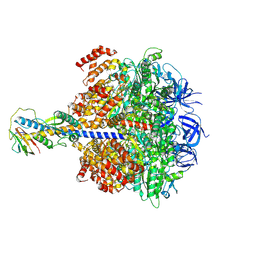 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
7U7N
 
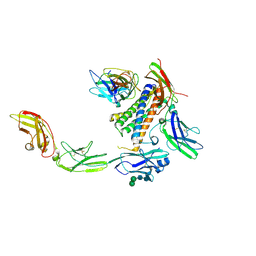 | | IL-27 quaternary receptor signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Caveney, N.A, Glassman, C.R, Jude, K.M, Tsutsumi, N, Garcia, K.C. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the IL-27 quaternary receptor signaling complex.
Elife, 11, 2022
|
|
9JVM
 
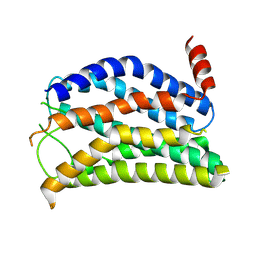 | |
6QKC
 
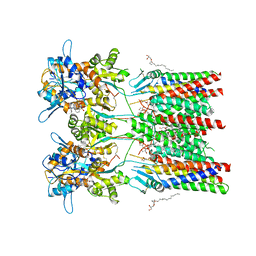 | | GluA1/2 In complex with auxiliary subunit gamma-8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 1, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.G. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma 8.
Science, 364, 2019
|
|
9JVG
 
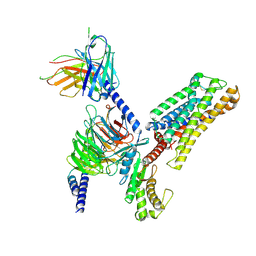 | | Cryo-EM structure of the mmGPR4-Gs complex in pH6.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | Deposit date: | 2024-10-09 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
9JVH
 
 | |
9DON
 
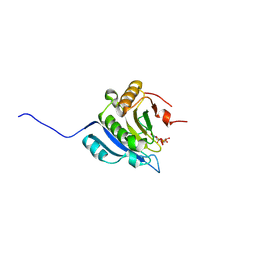 | | Crystal structure of eIF4e in complex with Compound 5-PA | | Descriptor: | 2-[3-[[2-azanyl-7-[(5-chloranyl-1-benzofuran-2-yl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]methyl]phenyl]ethylphosphonic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2024-09-19 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Second-Generation Cap Analogue Prodrugs for Targeting Aberrant Eukaryotic Translation Initiation Factor 4E Activity in Cancer.
Acs Med.Chem.Lett., 16, 2025
|
|
3ZPV
 
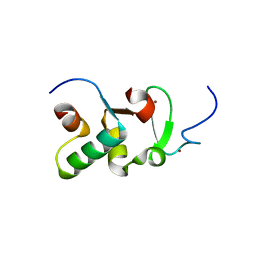 | | Crystal structure of Drosophila Pygo PHD finger in complex with Legless HD1 domain | | Descriptor: | PROTEIN BCL9 HOMOLOG, PROTEIN PYGOPUS, ZINC ION | | Authors: | Miller, T.C.R, Mieszczanek, J, Sanchez-Barrena, M.J, Rutherford, T.J, Fiedler, M, Bienz, M. | | Deposit date: | 2013-03-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Evolutionary Adaptation of the Fly Pygo Phd Finger Towards Recognizing Histone H3 Tail Methylated at Arginine 2
Structure, 21, 2013
|
|
8ZF4
 
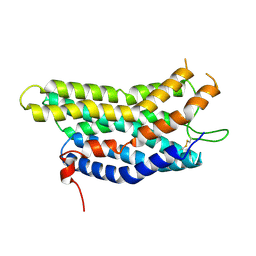 | |
8ZFB
 
 | |
8ZF7
 
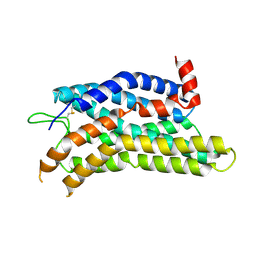 | |
8ZFC
 
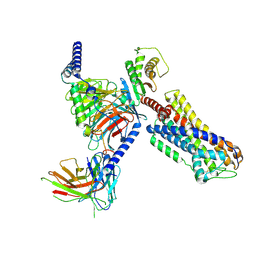 | | Cryo-EM structure of the mmGPR4-Gs complex in pH7.6 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZFE
 
 | |
8ZFA
 
 | | Cryo-EM structure of the xtGPR4-Gs complex in pH7.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
