2IZH
 
 | | STREPTAVIDIN-BIOTIN PH 10.44 I222 COMPLEX | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
8AZB
 
 | |
5FHN
 
 | |
8F7N
 
 | | Crystal structure of chemoreceptor McpZ ligand sensing domain | | Descriptor: | Methyl-accepting chemotaxis protein, YTTERBIUM (III) ION | | Authors: | Salar, S, Schubot, F.D. | | Deposit date: | 2022-11-19 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural analysis of the periplasmic domain of Sinorhizobium meliloti chemoreceptor McpZ reveals a novel fold and suggests a complex mechanism of transmembrane signaling.
Proteins, 91, 2023
|
|
8BRW
 
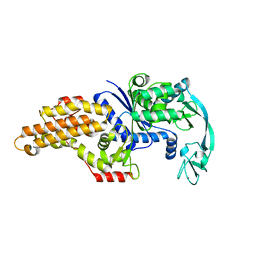 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C | | Descriptor: | Methionine--tRNA ligase, ZINC ION | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRX
 
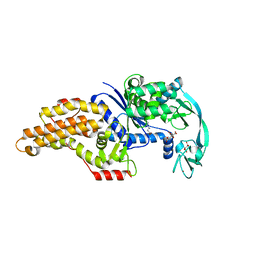 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C complexed with beta-3-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRV
 
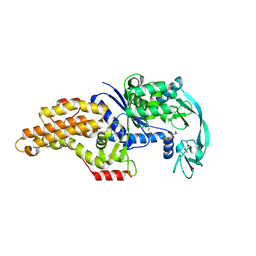 | | Escherichia coli methionyl-tRNA synthetase mutant L13M,I297C complexed with beta3-methionine. | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRU
 
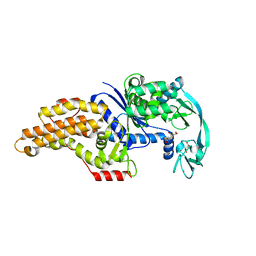 | | Escherichia coli methionyl-tRNA synthetase mutant L13M,I297C | | Descriptor: | CITRIC ACID, Methionine--tRNA ligase, ZINC ION | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
6FZ4
 
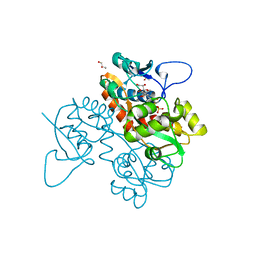 | | Structure of GluK1 ligand-binding domain in complex with N-(7-fluoro-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)-2-hydroxybenzamide at 1.85 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Mollerud, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N1-Substituted Quinoxaline-2,3-diones as Kainate Receptor Antagonists: X-ray Crystallography, Structure-Affinity Relationships, and in Vitro Pharmacology.
Acs Chem Neurosci, 10, 2019
|
|
8OOO
 
 | |
2RTK
 
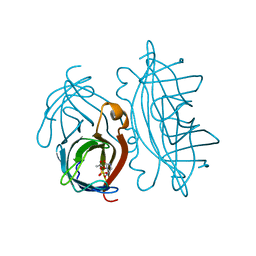 | |
9DB2
 
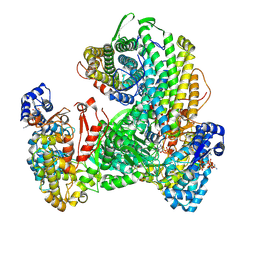 | | Class Ia ribonucleotide reductase with mechanism-based inhibitor N3CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 4-amino-1-{(3xi)-3-C-amino-2-deoxy-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-beta-D-threo-pentofuranosyl}pyrimidin-2(1H)-one, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Westmoreland, D.E, Drennan, C.L. | | Deposit date: | 2024-08-23 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | 2.6- angstrom resolution cryo-EM structure of a class Ia ribonucleotide reductase trapped with mechanism-based inhibitor N 3 CDP.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5VEY
 
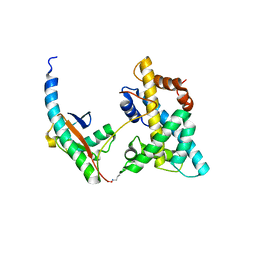 | | Solution NMR structure of histone H2A-H2B mono-ubiquitylated at H2A Lys15 in complex with RNF169 (653-708) | | Descriptor: | E3 ubiquitin-protein ligase RNF169, Histone H2B type 1-J,Histone H2A type 1-B/E, Polyubiquitin-B | | Authors: | Hu, Q, Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18.
Mol. Cell, 66, 2017
|
|
9F7X
 
 | |
9F7W
 
 | |
5ZYH
 
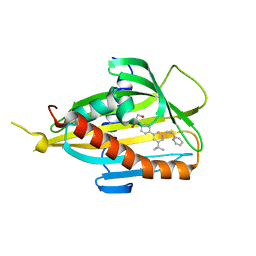 | | Crystal structure of CERT START domain in complex with compound E5 | | Descriptor: | 2-[4-[3-~{tert}-butyl-5-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYK
 
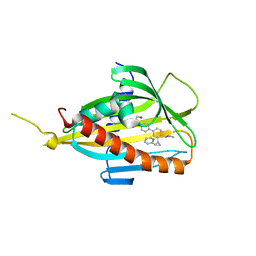 | | Crystal structure of CERT START domain in complex with compound E25 | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
4MLX
 
 | |
5ZYM
 
 | | Crystal structure of CERT START domain in complex with compound E25B | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5FHM
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-(aminomethyl)benzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at resolution 1.55 A resolution | | Descriptor: | (2~{S})-3-[5-[2-[[3-(aminomethyl)phenyl]methyl]-1,2,3,4-tetrazol-5-yl]-3-oxidanyl-1,2-oxazol-4-yl]-2-azanyl-propanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Al-musaed, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
1P1U
 
 | |
3OK9
 
 | | Crystal structure of wild-type HIV-1 protease with new oxatricyclic designed inhibitor GRL-0519A | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Probing Multidrug-Resistance and Protein-Ligand Interactions with Oxatricyclic Designed Ligands in HIV-1 Protease Inhibitors.
Chemmedchem, 5, 2010
|
|
8OR2
 
 | | CAND1-CUL1-RBX1-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8IFE
 
 | | Arbekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
