1R5U
 
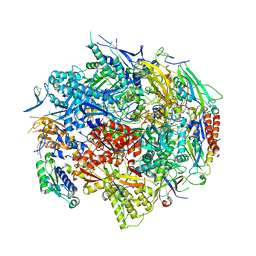 | | RNA POLYMERASE II TFIIB COMPLEX | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Bushnell, D.A, Westover, K.D, Davis, R, Kornberg, R.D. | | Deposit date: | 2003-10-13 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms.
Science, 303, 2004
|
|
5LCI
 
 | |
5LG5
 
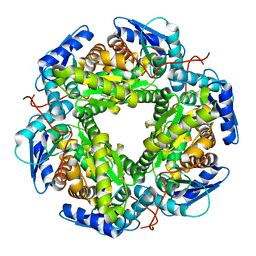 | | Crystal structure of allantoin racemase from Pseudomonas fluorescens AllR | | Descriptor: | Allantoin racemase | | Authors: | Cendron, l, Zanotti, G, Percudani, R, Ragazzina, I, Puggioni, V, Maccacaro, E, Liuzzi, A, Secchi, A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure and Function of a Microbial Allantoin Racemase Reveal the Origin and Conservation of a Catalytic Mechanism.
Biochemistry, 55, 2016
|
|
5LDG
 
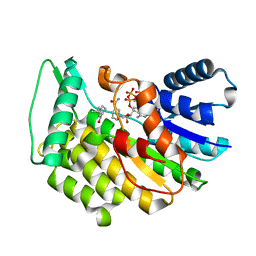 | | Isopiperitenone reductase from Mentha piperita in complex with Isopiperitenone and NADP | | Descriptor: | (-)-Isopiperitenone, (-)-isopiperitenone reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-06-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4HK6
 
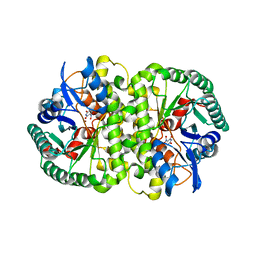 | |
8HKU
 
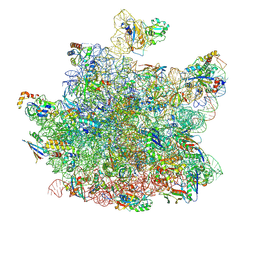 | |
2B5M
 
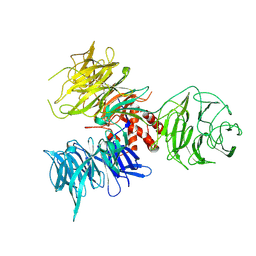 | | Crystal Structure of DDB1 | | Descriptor: | damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
8HKX
 
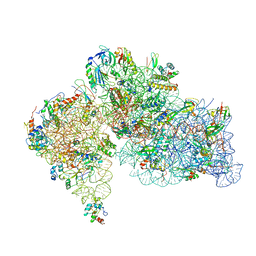 | |
1RGU
 
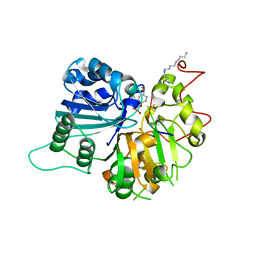 | | The crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTG | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*G)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
5LIT
 
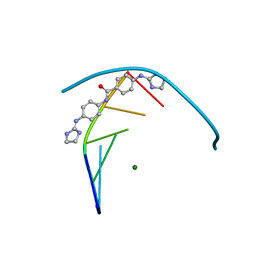 | | Structure of the DNA duplex d(AAATTT)2 with the potential antiparasitic drug 6XV at 1.25 A resolution | | Descriptor: | 4-((4,5-dihydro-1H-imidazol-2-yl)amino)-N-(4-((4,5-dihydro-1H-imidazol-2-yl)amino)phenyl)benzamide dihydrochloride, DNA (5'-D(*AP*AP*AP*TP*TP*T)-3'), MAGNESIUM ION | | Authors: | Millan, C.R, Dardonville, C, de Koning, H.P, Saperas, N, Lourdes Campos, J. | | Deposit date: | 2016-07-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.
Nucleic Acids Res., 45, 2017
|
|
2B3C
 
 | | SOLUTION STRUCTURE OF A BETA-NEUROTOXIN FROM THE NEW WORLD SCORPION CENTRUROIDES SCULPTURATUS EWING | | Descriptor: | PROTEIN (NEUROTOXIN CSE-I) | | Authors: | Jablonsky, M.J, Jackson, P.L, Trent, J.O, Watt, D.D, Krishna, N.R. | | Deposit date: | 1998-12-09 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a beta-neurotoxin from the New World scorpion Centruroides sculpturatus Ewing.
Biochem.Biophys.Res.Commun., 254, 1999
|
|
8HR2
 
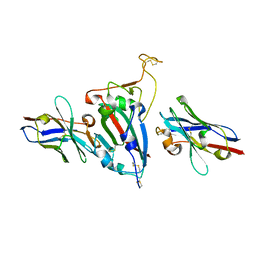 | | Ternary Crystal Complex Structure of RBD with NB1B5 and NB1C6 | | Descriptor: | NB1B5, NB1C6, Spike protein S1 | | Authors: | Sun, Z. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure basis of two nanobodies neutralizing SARS-CoV-2 Omicron variant by targeting ultra-conservative epitopes.
J.Struct.Biol., 215, 2023
|
|
8HKV
 
 | |
1RAG
 
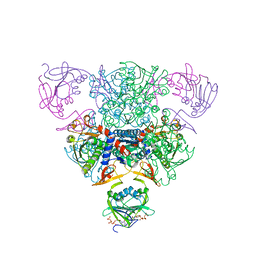 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
8HKZ
 
 | |
4WBI
 
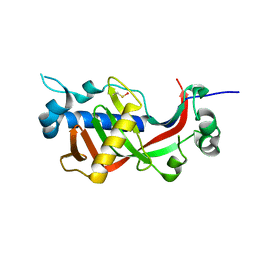 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutations H230Q and H309Q | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
1RP9
 
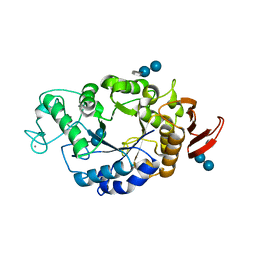 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
2B22
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4, SODIUM ION | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
4WCA
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H230Q, complexed with citrate | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
1RCK
 
 | |
5LQY
 
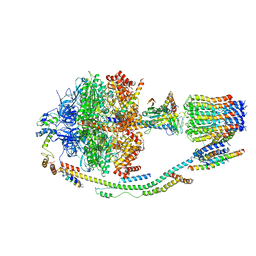 | | Structure of F-ATPase from Pichia angusta, in state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3F9V
 
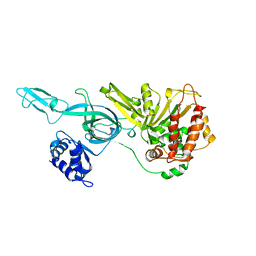 | | Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Chen, X.J, Brewster, A.S, Wang, G.G, Yu, X, Greenleaf, W, Tjajadi, M, Klein, M. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1TAR
 
 | |
8HL2
 
 | |
5LSE
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH Glu L212 replaced with Ala (CHAIN L, EL212W), Asp L213 replaced with ALA (Chain L, DL213A) AND LEU M215 REPLACED WITH ALA (CHAIN M, LM215A) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Jones, M.R. | | Deposit date: | 2016-08-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | On the mechanism of ubiquinone mediated photocurrent generation by a reaction center based photocathode.
Biochim.Biophys.Acta, 1857, 2016
|
|
