5R2D
 
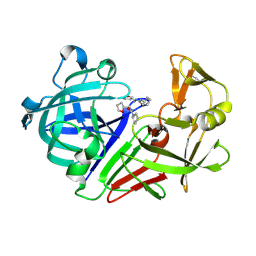 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H11, DMSO-free | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
3B60
 
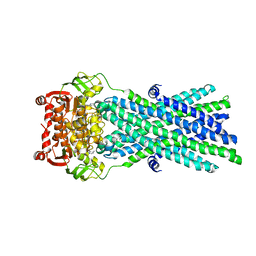 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP, higher resolution form | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3FBR
 
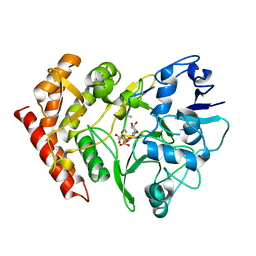 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
1MY4
 
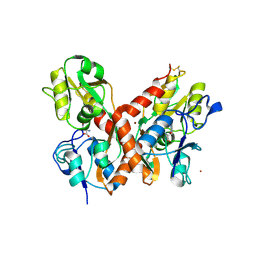 | | crystal structure of glutamate receptor ligand-binding core in complex with iodo-willardiine in the Zn crystal form | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
3B5Y
 
 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3FFB
 
 | | Gi-alpha-1 mutant in GDP bound form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Chauhan, R, Kapoor, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural evidence for a sequential release mechanism for activation of heterotrimeric g proteins.
J.Mol.Biol., 393, 2009
|
|
1Y2K
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 3,5-dimethyl-1-(3-nitro-phenyl)-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1,2-ETHANEDIOL, 3,5-DIMETHYL-1-(3-NITROPHENYL)-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
1QGZ
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 78 REPLACED BY ASP (L78D) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
1QH0
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 76 MUTATED BY ASP AND LEU 78 MUTATED BY ASP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
3C3Q
 
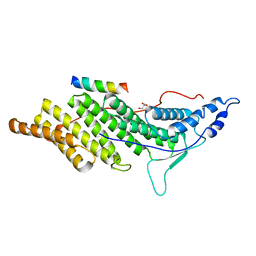 | | ALIX Bro1-domain:CHMIP4B co-crystal structure | | Descriptor: | Charged multivesicular body protein 4b peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1YTR
 
 | | NMR structure of plantaricin a in dpc micelles, 20 structures | | Descriptor: | Bacteriocin plantaricin A | | Authors: | Kristiansen, P.E, Fimland, G, Mantzilas, D, Nissen-Meyer, J. | | Deposit date: | 2005-02-11 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of the membrane-permeabilizing antimicrobial peptide pheromone plantaricin A
J.Biol.Chem., 280, 2005
|
|
2PUP
 
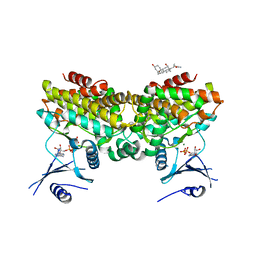 | |
1ZYD
 
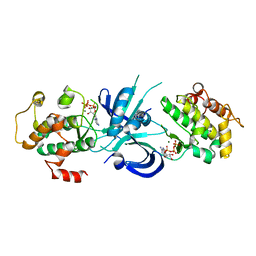 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: Wild-Type Complexed with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase GCN2 | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
2PU8
 
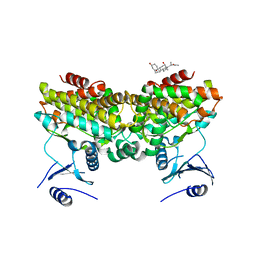 | |
2PUI
 
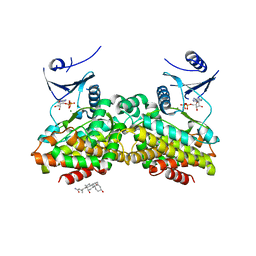 | |
5WOT
 
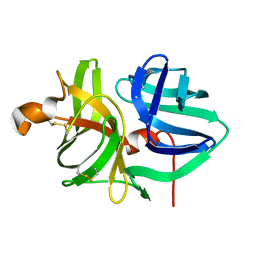 | |
3GU4
 
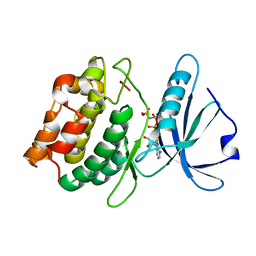 | |
5WOY
 
 | |
4XH5
 
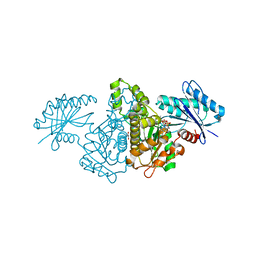 | | Crystal structure of Salmonella typhimurium propionate kinase A88G mutant, in complex with AMPPNP and propionate | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROPANOIC ACID, ... | | Authors: | Murthy, A.M, Mathivanan, S, Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-01-04 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structures of substrate- and nucleotide-bound propionate kinase from Salmonella typhimurium: substrate specificity and phosphate-transfer mechanism
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1DJC
 
 | |
7QI2
 
 | |
1DAW
 
 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF PROTEIN KINASE CK2 (ALPHA-SUBUNIT) AND MG-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN KINASE CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.G, Schomburg, D. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GTP plus water mimic ATP in the active site of protein kinase CK2.
Nat.Struct.Biol., 6, 1999
|
|
4XH4
 
 | | Crystal structure of Salmonella typhimurium propionate kinase A88V mutant, in complex with AMPPNP and propionate | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROPANOIC ACID, ... | | Authors: | Murthy, A.M.V, Mathivanan, S, Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-01-04 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of substrate- and nucleotide-bound propionate kinase from Salmonella typhimurium: substrate specificity and phosphate-transfer mechanism
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QXO
 
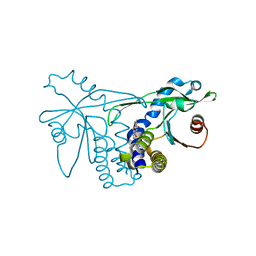 | | Crystal structure of hSTING(group2) in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
7M12
 
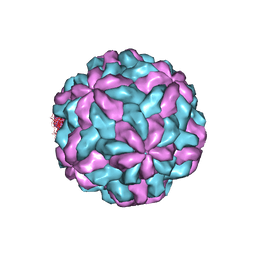 | | TVV2 capsid protein | | Descriptor: | Capsid protein | | Authors: | Zhou, H.Z, Stevens, A.W, Cui, Y.X, Muratore, K.A, Johnson, P.J. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.
Mbio, 12, 2021
|
|
