6TMK
 
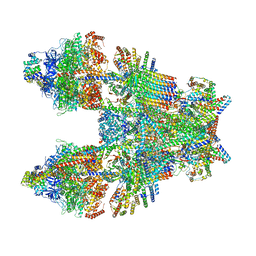 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, composite model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
8HH1
 
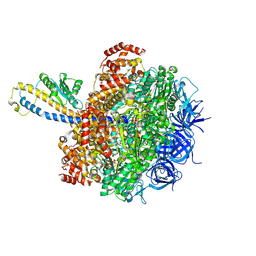 | | FoF1-ATPase from Bacillus PS3, 81 degrees, highATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH6
 
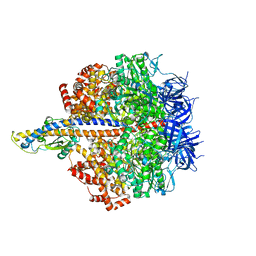 | | F1 domain of FoF1-ATPase from Bacillus PS3,step waiting,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH5
 
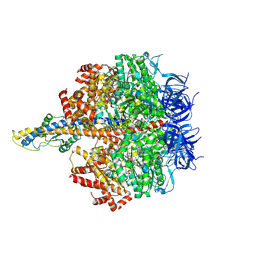 | | F1 domain of FoF1-ATPase from Bacillus PS3,120 degrees,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
6WLZ
 
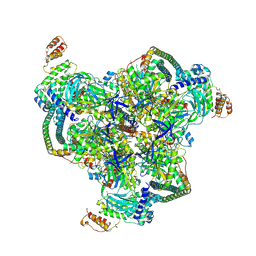 | | The V1 region of human V-ATPase in state 1 (focused refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SidK, V-type proton ATPase catalytic subunit A, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
8F2K
 
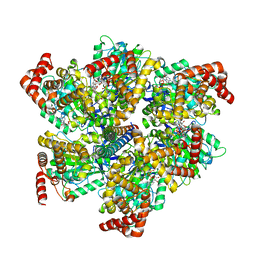 | |
1H8H
 
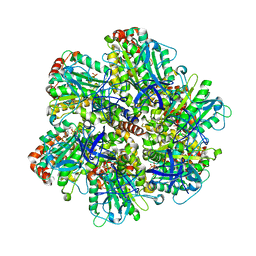 | | Bovine mitochondrial F1-ATPase crystallised in the presence of 5mm AMPPNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2001-02-06 | | Release date: | 2001-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Nucleotide Occupancy of Bovine Mitochondrial F(1)-ATPase are not Influenced by Crystallisation at High Concentrations of Nucleotide
FEBS Lett., 494, 2001
|
|
6RDJ
 
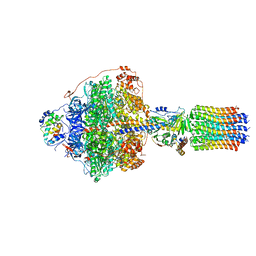 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
8G0A
 
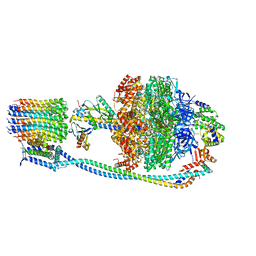 | |
8G0D
 
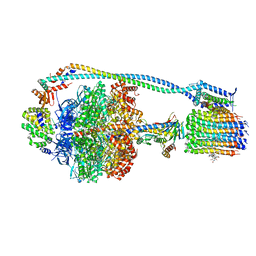 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
7NJO
 
 | | Mycobacterium smegmatis ATP synthase state 1e | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SPX
 
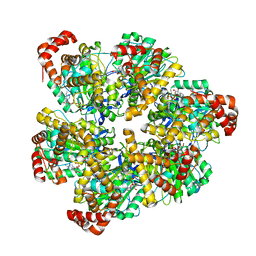 | | PS3 F1 Rotorless, high ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
3QG1
 
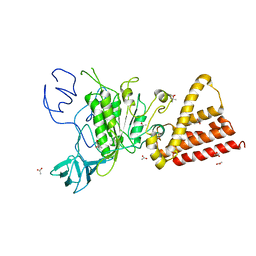 | | Crystal structure of P-loop G239A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, V-type ATP synthase alpha chain | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
7NKQ
 
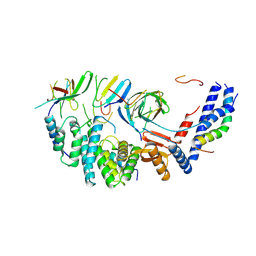 | | Mycobacterium smegmatis ATP synthase b-delta state 3 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5BN5
 
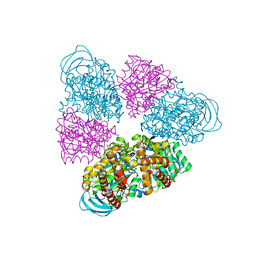 | | Structural basis for a unique ATP synthase core complex from Nanoarcheaum equitans | | Descriptor: | NEQ263, SULFATE ION, V-type ATP synthase alpha chain | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
6RD9
 
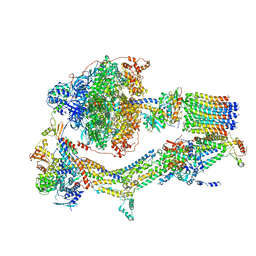 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | Authors: | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | Deposit date: | 1998-04-30 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
6RDH
 
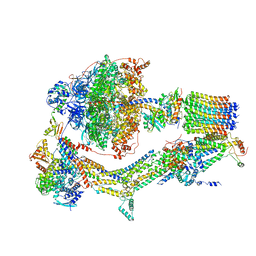 | | CryoEM structure of Polytomella F-ATP synthase, Rotary substate 1A, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RED
 
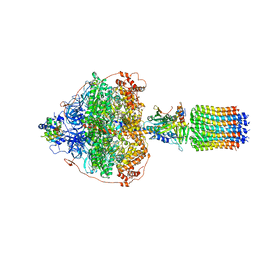 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
7YRY
 
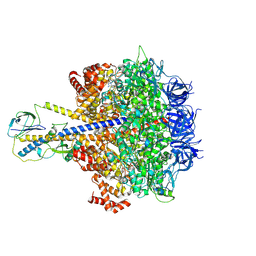 | | F1-ATPase of Acinetobacter baumannii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Grueber, G. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic insights of an up and down conformation of the Acinetobacter baumannii F 1 -ATPase subunit epsilon and deciphering the residues critical for ATP hydrolysis inhibition and ATP synthesis.
Faseb J., 37, 2023
|
|
8GXX
 
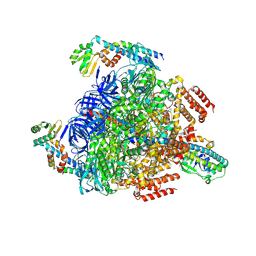 | | 3 nucleotide-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
7VAP
 
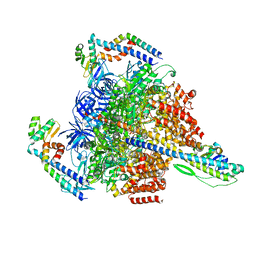 | | V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state2-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
3OEH
 
 | | Structure of four mutant forms of yeast F1 ATPase: beta-V279F | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
7VAN
 
 | | V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state2-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAS
 
 | | V1EG domain of V/A-ATPase from Thermus thermophilus at low ATP concentration, state1-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
