2MOI
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MOL
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
3HIX
 
 | | Crystal Structure of the Rhodanese_3 like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437i | | Descriptor: | Alr3790 protein, MANGANESE (II) ION | | Authors: | Vorobiev, S, Chen, Y, Forouhar, F, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the Rhodanese_3 like domain from Anabaena sp Alr3790 protein.
To be Published
|
|
3IPO
 
 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IPP
 
 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IWH
 
 | | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus | | Descriptor: | BETA-MERCAPTOETHANOL, Rhodanese-like domain protein | | Authors: | Kim, Y, Chruszcz, M, Minor, W, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus
To be Published
|
|
3ILM
 
 | | Crystal Structure of the Alr3790 protein from Anabaena sp. Northeast Structural Genomics Consortium Target NsR437h | | Descriptor: | Alr3790 protein, MANGANESE (II) ION | | Authors: | Vorobiev, S, Chen, Y, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Crystal Structure of the Alr3790 protein from Anabaena sp.
To be Published
|
|
3K9R
 
 | | X-ray structure of the Rhodanese-like domain of the Alr3790 protein from Anabaena sp. Northeast Structural Genomics Consortium Target NsR437c. | | Descriptor: | Alr3790 protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of the Rhodanese-like domain of the Alr3790 protein from Anabaena sp.
To be Published
|
|
3I2V
 
 | | Crystal structure of human MOCS3 rhodanese-like domain | | Descriptor: | Adenylyltransferase and sulfurtransferase MOCS3 | | Authors: | Bacik, J.P, Walker, J.R, Lopez, L, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the human MOCS3 rhodanese-like domain
To be Published
|
|
3HWI
 
 | |
3I3U
 
 | | Crystal structure of lp_1913 protein from lactobacillus plantarum, northeast structural genomics Consortium target lpr140a | | Descriptor: | uncharacterized protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of lp_1913 protein from lactobacillus plantarum, northeast structural genomics Consortium target lpr140a
To be Published
|
|
3HZU
 
 | |
4OCG
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase F161A Mutant | | Descriptor: | COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Lee, K.-H, Sazinsky, M.H, Crane, E.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Characterization of the mechanism of the NADH-dependent polysulfide reductase (Npsr) from Shewanella loihica PV-4: Formation of a productive NADH-enzyme complex and its role in the general mechanism of NADH and FAD-dependent enzymes.
Biochim.Biophys.Acta, 1844, 2014
|
|
4F67
 
 | | Three dimensional structure of the double mutant of UPF0176 protein lpg2838 from Legionella pneumophila at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target LgR82 | | Descriptor: | UPF0176 protein lpg2838 | | Authors: | Kuzin, A, Neely, H, Street, L, Odukwe, N, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-14 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Three dimensional structure of the double mutant of UPF0176 protein lpg2838 from Legionella pneumophila at the resolution 1.8A, Northeast
Structural Genomics Consortium (NESG) Target LgR82
To be Published
|
|
8A56
 
 | | Coenzyme A-persulfide reductase (CoAPR) from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Coenzyme A-persulfide reductase, ... | | Authors: | Costa, S.S, Walsh, B.J, Giedroc, D.P, Brito, J.A. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Metabolic and Structural Insights into Hydrogen Sulfide Mis-Regulation in Enterococcus faecalis.
Antioxidants, 11, 2022
|
|
6YUB
 
 | | Crystal structure of Uba4 from Chaetomium thermophilum | | Descriptor: | Adenylyltransferase and sulfurtransferase uba4, ZINC ION | | Authors: | Grudnik, P, Pabis, M, Ethiraju Ravichandran, K, Glatt, S. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation.
Embo J., 39, 2020
|
|
5VE4
 
 | | Crystal structure of persulfide dioxygenase-rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans | | Descriptor: | BpPRF, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5VE5
 
 | | Crystal structure of persulfide dioxygenase rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans in complex with glutathione | | Descriptor: | BpPRF, CHLORIDE ION, FE (III) ION, ... | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5VE3
 
 | | Crystal structure of wild-type persulfide dioxygenase-rhodanese fusion protein from Burkholderia phytofirmans | | Descriptor: | BpPRF, FE (III) ION | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
3R2U
 
 | | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase from Staphylococcus aureus subsp. aureus COL | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase Family Protein from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3TP9
 
 | | Crystal structure of Alicyclobacillus acidocaldarius protein with beta-lactamase and rhodanese domains | | Descriptor: | BETA-LACTAMASE and RHODANESE DOMAIN PROTEIN, ZINC ION | | Authors: | Michalska, K, Chhor, G, Mandel, M.E, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Alicyclobacillus acidocaldarius protein with beta-lactamase and rhodanese domains
To be Published
|
|
2GWF
 
 | | Structure of a USP8-NRDP1 complex | | Descriptor: | RING finger protein 41, Ubiquitin carboxyl-terminal hydrolase 8 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
1OKG
 
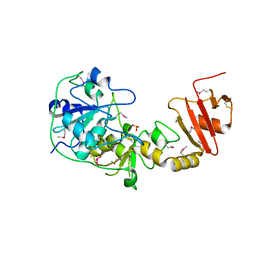 | | 3-mercaptopyruvate sulfurtransferase from Leishmania major | | Descriptor: | CALCIUM ION, POSSIBLE 3-MERCAPTOPYRUVATE SULFURTRANSFERASE, SULFITE ION | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2003-07-24 | | Release date: | 2003-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Leishmania Major 3-Mercaptopyruvate Sulfurtransferase: A Three-Domain Architecture with a Serine Protease-Like Triad at the Active Site
J.Biol.Chem., 278, 2003
|
|
3NTD
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C531S Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Warner, M.D, Lukose, V, Lee, K.H, Crane, E.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NTA
 
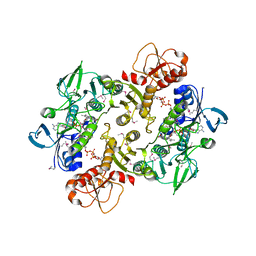 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
