3MSD
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
7NCV
 
 | |
1PYF
 
 | |
3MPN
 
 | | F177R1 mutant of LeuT | | Descriptor: | CHLORIDE ION, LEUCINE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ... | | Authors: | Kroncke, B.M. | | Deposit date: | 2010-04-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural origins of nitroxide side chain dynamics on membrane protein alpha-helical sites.
Biochemistry, 49, 2010
|
|
7NJF
 
 | | Hen egg white lysozyme (HEWL) grown inside HARE serial crystallography chip | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NKF
 
 | | Hen egg white lysozyme (HEWL) Grown inside (Not centrifuged) HARE serial crystallography chip. | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2I2S
 
 | | Crystal Structure of the porcine CRW-8 rotavirus VP8* carbohydrate-recognising domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, ... | | Authors: | Blanchard, H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into Host Cell Carbohydrate-recognition by Human and Porcine Rotavirus from Crystal Structures of the Virion Spike Associated Carbohydrate-binding Domain (VP8*)
J.Mol.Biol., 367, 2007
|
|
7NEI
 
 | |
2IAG
 
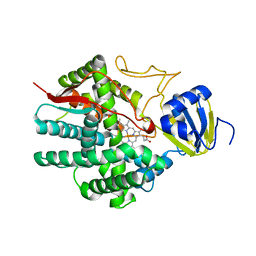 | | Crystal structure of human prostacyclin synthase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostacyclin synthase, SODIUM ION | | Authors: | Chiang, C.-W, Yeh, H.-C, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2006-09-08 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Prostacyclin Synthase
J.Mol.Biol., 364, 2006
|
|
1OB7
 
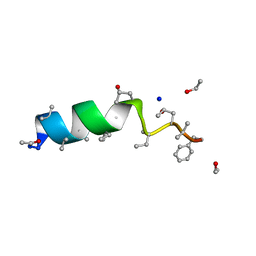 | | Cephaibol C | | Descriptor: | CEPHAIBOL C, ETHANOL, SODIUM ION | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
3NKO
 
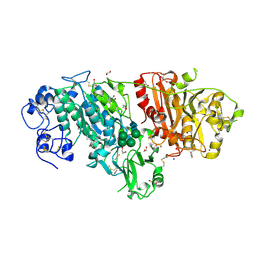 | | Crystal structure of mouse autotaxin in complex with 16:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
1O4Y
 
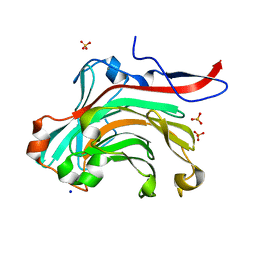 | | THE THREE-DIMENSIONAL STRUCTURE OF BETA-AGARASE A FROM ZOBELLIA GALACTANIVORANS | | Descriptor: | CALCIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Allouch, J, Jam, M, Helbert, W, Barbeyron, T, Kloareg, B, Henrissat, B, Czjzek, M. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Three-dimensional Structures of Two {beta}-Agarases.
J.Biol.Chem., 278, 2003
|
|
7NOW
 
 | |
4DXO
 
 | |
7NF5
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NEU
 
 | | Inhibitor Complex with Thrombin Activatable Fibrinolysis Inhibitor (TAFIa) | | Descriptor: | (1R,3S)-3-(4-ammoniobutyl)-1-(4-fluoro-2-(1-methyl-1H-imidazol-5-yl)benzyl)-1,4-azaphosphinan-1-ium-3-carboxylate 4,4-dioxide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brown, D.G, Schaffner, A.P, Vuillard, L.M, Gloanec, P, Raimbauld, E. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphinanes and Azaphosphinanes as Potent and Selective Inhibitors of Activated Thrombin-Activatable Fibrinolysis Inhibitor (TAFIa).
J.Med.Chem., 64, 2021
|
|
3NKP
 
 | | Crystal structure of mouse autotaxin in complex with 18:1-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
7NCD
 
 | | Glutathione-S-transferase GliG mutant N27D | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCP
 
 | | Glutathione-S-transferase GliG mutant K127A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG, SODIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NDH
 
 | | Crystal structure of ZC3H12C PIN domain | | Descriptor: | 1,2-ETHANEDIOL, Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDJ
 
 | |
7NDI
 
 | | Crystal structure of ZC3H12C PIN domain with Mg2+ Ion | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable ribonuclease ZC3H12C, ... | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDK
 
 | | Crystal structure of ZC3H12C PIN catalytic mutant | | Descriptor: | Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
3NKR
 
 | | Crystal structure of mouse autotaxin in complex with 22:6-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (4Z,7E,10E,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
4E0S
 
 | | Crystal Structure of C5b-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C5, ... | | Authors: | Aleshin, A.E, Stec, B, DiScipio, R, Liddington, R.C. | | Deposit date: | 2012-03-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.21 Å) | | Cite: | Crystal structure of c5b-6 suggests structural basis for priming assembly of the membrane attack complex.
J.Biol.Chem., 287, 2012
|
|
