1GB2
 
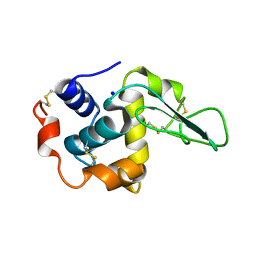 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GB9
 
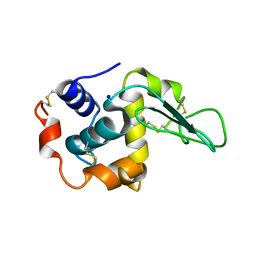 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GB5
 
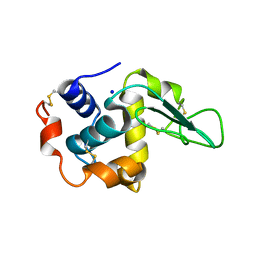 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GF0
 
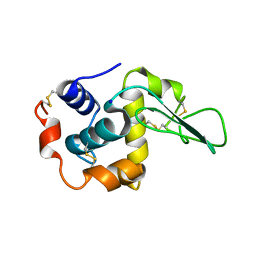 | | BURIED POLAR MUTANT HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of polar groups in the interior of a protein to the conformational stability.
Biochemistry, 40, 2001
|
|
1GF6
 
 | | BURIED POLAR MUTANT HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of polar groups in the interior of a protein to the conformational stability.
Biochemistry, 40, 2001
|
|
1GF8
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFE
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
7X0T
 
 | |
7X0Z
 
 | |
7XE6
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH7 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XEA
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 40%, and then backsoaking | | Descriptor: | CHLORIDE ION, Endolysin, GLYCEROL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7X07
 
 | | Cryo-EM structure of human ABCD1 in the presence of C26:0 | | Descriptor: | ATP-binding cassette sub-family D member 1, hexacosanoic acid | | Authors: | Chao, X, Li-Na, J, Lin, T. | | Deposit date: | 2022-02-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insights into substrate recognition and translocation of human peroxisomal ABC transporter ALDP.
Signal Transduct Target Ther, 8, 2023
|
|
7XE9
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 20% | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Endolysin, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE5
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH4 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7JXN
 
 | |
7XBE
 
 | |
7XEC
 
 | |
7X1W
 
 | |
1Z3Z
 
 | | The crystal structure of a DGD mutant: Q52A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fogle, E.J, Liu, W, Toney, M.D. | | Deposit date: | 2005-03-14 | | Release date: | 2006-01-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of q52 in catalysis of decarboxylation and transamination in dialkylglycine decarboxylase.
Biochemistry, 44, 2005
|
|
7XFT
 
 | | MucP PDZ2 domain | | Descriptor: | Putative zinc metalloprotease PA3649, SODIUM ION, VALINE | | Authors: | Lou, X, Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-04-02 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | MucP PDZ2 domain
To Be Published
|
|
1GDW
 
 | |
1GFV
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
7K16
 
 | | Tamana Bat Virus xrRNA1 | | Descriptor: | MAGNESIUM ION, RNA (51-MER), SODIUM ION | | Authors: | Jones, R.A, Kieft, J.S. | | Deposit date: | 2020-09-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different tertiary interactions create the same important 3D features in a distinct flavivirus xrRNA.
Rna, 27, 2021
|
|
1GJ4
 
 | | SELECTIVITY AT S1, H2O DISPLACEMENT, UPA, TPA, SER190/ALA190 PROTEASE, STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1ZKO
 
 | |
