1AGB
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGRKKYKL-3R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGRKKYKL - 3R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
3E4U
 
 | | Crystal Structure of the Wild-Type Human BCL6 BTB/POZ Domain | | Descriptor: | B-cell lymphoma 6 protein | | Authors: | Stead, M.A, Rosbrook, G.O, Hadden, J.M, Trinh, C.H, Carr, S.B, Wright, S.C. | | Deposit date: | 2008-08-12 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the wild-type human BCL6 POZ domain.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1AGC
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYQL-7Q MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYQL - 7Q MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGF
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKRYKL-5R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKRYKL - 5R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGD
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYKL-INDEX PEPTIDE) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYKL - INDEX PEPTIDE) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGE
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYRL-7R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYRL - 7R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
9FZM
 
 | |
6UAN
 
 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
4XV9
 
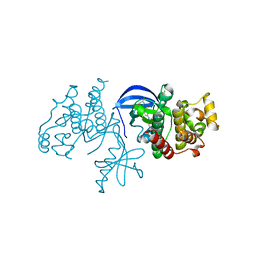 | | B-Raf Kinase domain in complex with PLX5568 | | Descriptor: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}-4-(trifluoromethyl)benzenesulfonamide, SULFATE ION, Serine/threonine-protein kinase B-raf | | Authors: | zhang, Y, zhang, c, wang, w. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV1
 
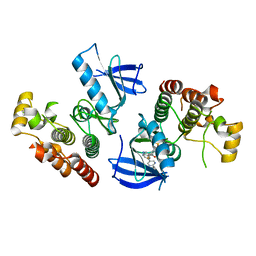 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7904 | | Descriptor: | N'-(3-{[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
2KIX
 
 | |
3LCZ
 
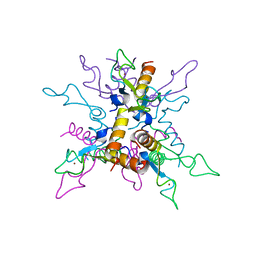 | | B.licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
6CNV
 
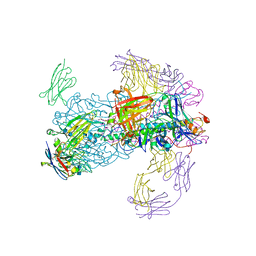 | | INFLUENZA B/BRISBANE HEMAGGLUTININ FAB CR9115 SD84H COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR9114 Fab heavy chain, ... | | Authors: | Luo, J, Obmolova, G. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
3II5
 
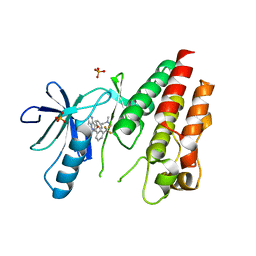 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4G9R
 
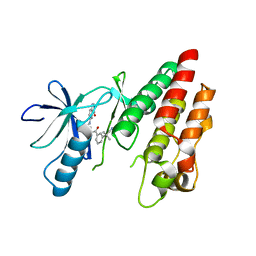 | | B-Raf V600E Kinase Domain Bound to a Type II Dihydroquinazoline Inhibitor | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-{4-methyl-3-[(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Sturgis, H.L. | | Deposit date: | 2012-07-24 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pyrazolopyridine inhibitors of B-Raf(V600E). Part 4: Rational design and kinase selectivity profile of cell potent type II inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5FD2
 
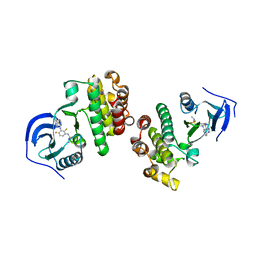 | |
1VIB
 
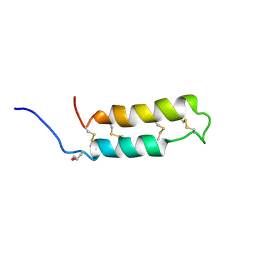 | | NMR SOLUTION STRUCTURE OF THE NEUROTOXIN B-IV, 20 STRUCTURES | | Descriptor: | NEUROTOXIN B-IV | | Authors: | Barnham, K.J, Dyke, T.R, Kem, W.R, Norton, R.S. | | Deposit date: | 1996-11-25 | | Release date: | 1997-05-15 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Structure of neurotoxin B-IV from the marine worm Cerebratulus lacteus: a helical hairpin cross-linked by disulphide bonding.
J.Mol.Biol., 268, 1997
|
|
3Q96
 
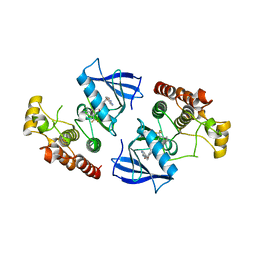 | | B-Raf kinase domain in complex with a tetrahydronaphthalene inhibitor | | Descriptor: | (2S)-N-[3-(2-aminopropan-2-yl)-5-(trifluoromethyl)phenyl]-7-[(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-4-yl)oxy]-1,2,3,4-tetrahydronaphthalene-2-carboxamide, Serine/threonine-protein kinase B-raf | | Authors: | Sintchak, M.D, Aertgeerts, K, Yano, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Design and optimization of potent and orally bioavailable tetrahydronaphthalene raf inhibitors.
J.Med.Chem., 54, 2011
|
|
1DCV
 
 | |
4UZH
 
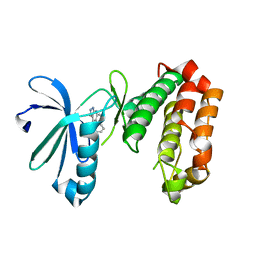 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | (4S)-4-(2-fluorophenyl)-2,4,6,7,8,9-hexahydro-5H-pyrazolo[3,4-b][1,7]naphthyridin-5-one, AURORA 2 KINASE DOMAIN | | Authors: | Pouzieux, S, Maignan, S, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4URO
 
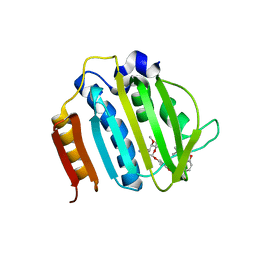 | | Crystal Structure of Staph GyraseB 24kDa in complex with Novobiocin | | Descriptor: | DNA GYRASE SUBUNIT B, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4UZD
 
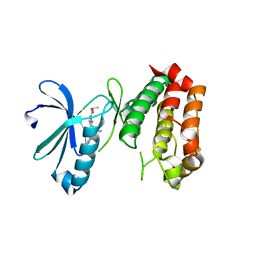 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[3-(1H-benzimidazol-2-yloxy)phenyl]-8-oxo-4,5,6,7,8,9-hexahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Pouzieux, S, Delarbre, L, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
7O1P
 
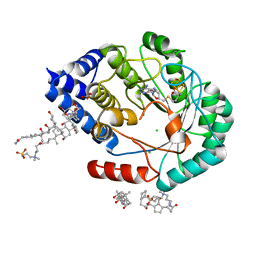 | | [FeFe]-hydrogenase maturase HydE from T. Maritima (C-ter stretch absent) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1T
 
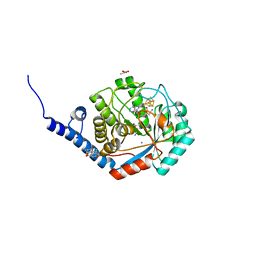 | | Fe(CO)2CNCl species bound [HydE from T. Maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7MYJ
 
 | | Structure of full length human AMPK (a2b1g1) in complex with a small molecule activator MSG011 | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Ovens, A.J, Gee, Y.S, Ling, N.X.Y, Waters, N.J, Yu, D, Scott, J.W, Parker, M.W, Hoffman, N.J, Kemp, B.E, Baell, J.B, Oakhill, J.S, Langendorf, C.G. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-function analysis of the AMPK activator SC4 and identification of a potent pan AMPK activator.
Biochem.J., 479, 2022
|
|
