7NUN
 
 | | Rhinovirus 14 ICAM-1 virion-like particle at pH 6.2 | | Descriptor: | Genome polyprotein, Octanucleotide | | Authors: | Hrebik, D, Plevka, P. | | Deposit date: | 2021-03-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8BSK
 
 | | Human GLS in complex with compound 3 | | Descriptor: | Glutaminase kidney isoform, mitochondrial 65 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide), ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
6GPA
 
 | | Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose | | Descriptor: | Arabinogalactan endo-beta-1,4-galactanase, beta-D-galactopyranose | | Authors: | Hekelaar, J, Boger, M, Leeuwen van, S.S, Lammerts van Bueren, A, Dijkhuizen, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of a family GH53 beta-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides.
J. Struct. Biol., 205, 2019
|
|
6GP5
 
 | | Beta-1,4-galactanase from Bacteroides thetaiotaomicron | | Descriptor: | Arabinogalactan endo-beta-1,4-galactanase | | Authors: | Hekelaar, J, Boger, M, Leeuwen van, S.S, Lammerts van Bueren, A, Dijkhuizen, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and functional characterization of a family GH53 beta-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides.
J. Struct. Biol., 205, 2019
|
|
6XX7
 
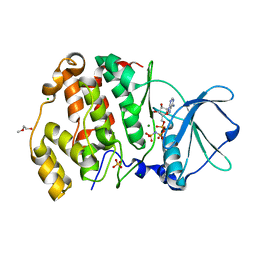 | | Arabidopsis thaliana Casein Kinase 2 (CK2) alpha-1 crystal in complex with ANP | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha-1, MAGNESIUM ION, ... | | Authors: | Demulder, M, De Veylder, L, Loris, R. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Arabidopsis thaliana casein kinase 2 alpha 1.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8OS1
 
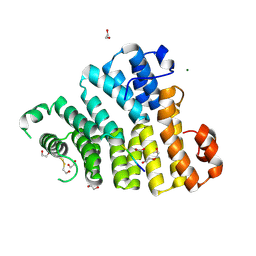 | | X-ray structure of the Peroxisomal Targeting Signal 1 (PTS1) of Trypanosoma Cruzi PEX5 in complex with the PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Napolitano, V, Blat, A, Popowicz, G.M, Dubin, G. | | Deposit date: | 2023-04-17 | | Release date: | 2024-10-02 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dynamics of the TPR domain of the peroxisomal cargo receptor Pex5 in Trypanosoma.
Int.J.Biol.Macromol., 280, 2024
|
|
6XX6
 
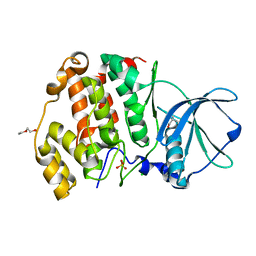 | | Arabidopsis thaliana Casein Kinase 2 (CK2) alpha-1 crystal form I | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha-1, PIVALIC ACID, ... | | Authors: | Demulder, M, De Veylder, L, Loris, R. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84919691 Å) | | Cite: | Crystal structure of Arabidopsis thaliana casein kinase 2 alpha 1.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7N6X
 
 | |
9DV9
 
 | |
6ISN
 
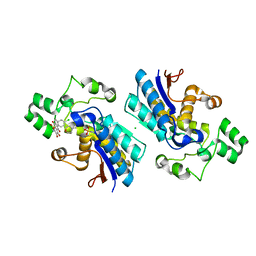 | | Phosphoglycerate mutase 1 complexed with a small molecule inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-chloro-N-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracen-2-yl)benzene-1-sulfonamide, CHLORIDE ION, ... | | Authors: | Jiang, L.L, Zhou, L, Huang, K. | | Deposit date: | 2018-11-17 | | Release date: | 2019-12-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a small molecule inhibitor
To Be Published
|
|
5CTE
 
 | |
9DV8
 
 | |
6E6X
 
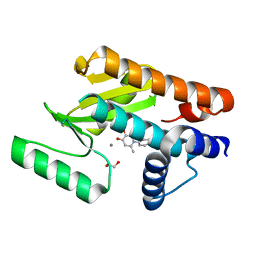 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 1-(4-(1H-tetrazol-5-yl)phenyl)-5-hydroxy-2-methylpyridin-4(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 5-hydroxy-2-methyl-1-[4-(1H-tetrazol-5-yl)phenyl]pyridin-4(1H)-one, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
7AX3
 
 | |
5L78
 
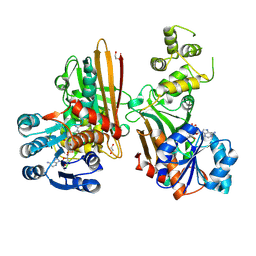 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in NAD+ bound form) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Pena, I.A, Rembeza, E, Strain-Damerell, C, Chalk, R, Borkowska, O, Goubin, S, Velupillai, S, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Arruda, P, Yue, W.W. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in NAD+ bound form)
To Be Published
|
|
5LYM
 
 | |
9BFW
 
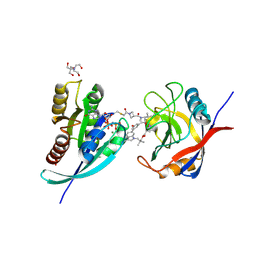 | | Tri-complex of Compound-4, KRAS G12C, and CypA | | Descriptor: | 1-acetyl-N-[(2S)-1-{[(1M,8S,10R,14S,20S)-22-cyano-4-hydroxy-18,18-dimethyl-9,15-dioxo-16-oxa-10,20,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,21,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methylazetidine-3-carboxamide (non-preferred name), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Elironrasib (RMC-6291), a Potent and Orally Bioavailable, RAS(ON) G12C-Selective, Covalent Tricomplex Inhibitor for the Treatment of Patients with RAS G12C-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
9BFX
 
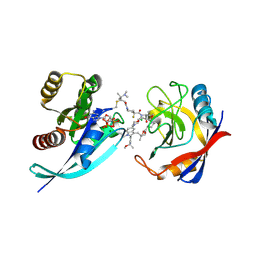 | | Tri-complex of Elironrasib (RMC-6291), KRAS G12C, and CypA | | Descriptor: | 1-[(2E)-4-(dimethylamino)-4-methylpent-2-enoyl]-N-[(2R)-1-{[(2S,6S,8S,10R,14S,21M)-22-ethyl-21-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-5,16-dioxa-2,10,22,28-tetraazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),20,23,26-tetraen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-4-fluoro-N-methylpiperidine-4-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of Elironrasib (RMC-6291), a Potent and Orally Bioavailable, RAS(ON) G12C-Selective, Covalent Tricomplex Inhibitor for the Treatment of Patients with RAS G12C-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
7AXY
 
 | |
9ARP
 
 | |
9ARN
 
 | |
7B3I
 
 | | Notum complex with ARUK3003776 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-chlorophenyl)sulfanyl-1$l^{4},2,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
9K0G
 
 | | Structure of Human HDAC2 in complex with inhibitor N-(2-aminophenyl)-4-(1-((phenylsulfonyl)methyl)-1H-1,2,3-triazol-4-yl)benzamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tojo, T, Itoh, Y, Kurohara, T, Li, Y, Singh, R, Narozny, R, Wiel, A, Miyake, Y, Yamashita, Y, Kusakabe, K, Uchida, S, Suzuki, T. | | Deposit date: | 2024-10-15 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of an HDAC2-selective inhibitor based on enzyme-inhibitor binding thermodynamics and kinetics, and its potential as a therapeutic drug for neurological disorders
To Be Published
|
|
5KVH
 
 | | Crystal structure of human apoptosis-inducing factor with W196A mutation | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
8PFE
 
 | |
