9DH6
 
 | |
9DH7
 
 | |
6RVK
 
 | | Crystal structure of hCA II in complex with Urea, N-(1,3-dihydro-1-hydroxy-2,1-benzoxaborol-6-yl)-N'-(phenylmethyl)- | | Descriptor: | 1-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-4-yl]-3-(phenylmethyl)urea, Carbonic anhydrase 2, ZINC ION | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2019-05-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring benzoxaborole derivatives as carbonic anhydrase inhibitors: a structural and computational analysis reveals their conformational variability as a tool to increase enzyme selectivity.
J Enzyme Inhib Med Chem, 34, 2019
|
|
5LUC
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.8 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Borri Voltattorni, C, Montioli, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
9DH9
 
 | |
6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
9DH8
 
 | |
6RHJ
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-(4-benzyl-1,4-diazepane-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[[4-(phenylmethyl)-1,4-diazepan-1-yl]carbonyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
9E7D
 
 | |
7ZZW
 
 | | Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZU
 
 | | Inhibitory Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R},4~{S})-4-phenylpyrrolidin-2-yl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
5YKT
 
 | |
8HY3
 
 | |
7YNO
 
 | |
5NWW
 
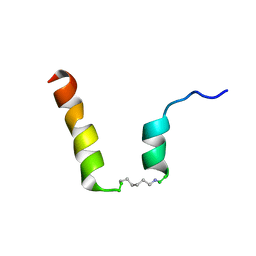 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
9E10
 
 | | Dimeric motor domains from phi dynein-1 under Lis1 condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Yang, J, Zhang, K. | | Deposit date: | 2024-10-21 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Nde1 promotes Lis1 binding to full-length autoinhibited human dynein 1.
Nat.Chem.Biol., 2025
|
|
9KCA
 
 | | Cryo-EM structure of docked mouse bestrophin-1 in a closed state | | Descriptor: | Bestrophin-1,Soluble cytochrome b562, CALCIUM ION, CHLORIDE ION | | Authors: | Lim, H.H, Kim, K.W, Ko, A. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM structures of mouse bestrophin 1 channel in closed and partially open conformations.
Mol.Cells, 48, 2025
|
|
9KC9
 
 | | Cryo-EM structure of docked mouse bestrophin-1 in a partial open state | | Descriptor: | Bestrophin-1,Soluble cytochrome b562, CALCIUM ION, CHLORIDE ION | | Authors: | Lim, H.H, Kim, K.W, Ko, A. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of mouse bestrophin 1 channel in closed and partially open conformations.
Mol.Cells, 48, 2025
|
|
9KP6
 
 | | Cryo-EM structure of mouse bestrophin-1 in a closed state | | Descriptor: | Bestrophin-1,Soluble cytochrome b562, CALCIUM ION, CHLORIDE ION | | Authors: | Lim, H.H, Kim, K.W, Ko, A. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM structures of mouse bestrophin 1 channel in closed and partially open conformations.
Mol.Cells, 48, 2025
|
|
8PPC
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with L-chiro-inositol 2,3,5-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-trisphosphate 3-kinase A, L-chiro-inositol 2,3,5-trisphosphate, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
5OKC
 
 | |
7CKG
 
 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | Descriptor: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
6FY0
 
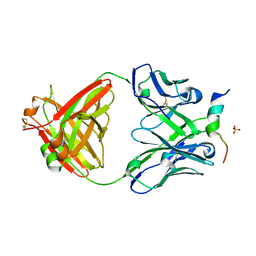 | | Crystal structure of a V2-directed, RV144 vaccine-like antibody from HIV-1 infection, CAP228-16H, bound to a heterologous V2 peptide | | Descriptor: | CAP228-16H Heavy Chain, CAP228-16H Light Chain, CAP45 V2 peptide, ... | | Authors: | Wibmer, C.K, Fernandes, M, Vijayakumar, B, Dirr, H.W, Sayed, Y, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | V2-Directed Vaccine-like Antibodies from HIV-1 Infection Identify an Additional K169-Binding Light Chain Motif with Broad ADCC Activity.
Cell Rep, 25, 2018
|
|
5IDL
 
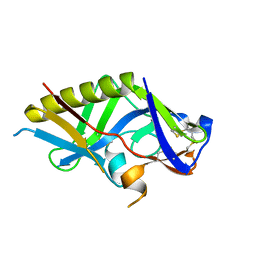 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Authors: | Julien, J.P, Ereno-Orbea, J, Jardine, J.G, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.
Science, 351, 2016
|
|
1QMS
 
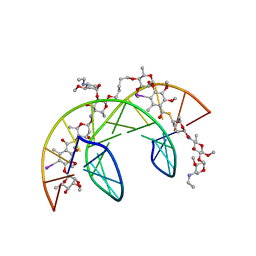 | | Head-to-Tail Dimer of Calicheamicin gamma-1-I Oligosaccharide Bound to DNA Duplex, NMR, 9 Structures | | Descriptor: | CALICHEAMICIN GAMMA-1-OLIGOSACCHARIDE, DNA (5'-D(*GP*CP*AP*CP*CP*TP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*AP*GP*GP*TP*GP*C)-3'), ... | | Authors: | Bifulco, G, Galeone, A, Nicolaou, K.C, Chazin, W.J, Gomez-Paloma, L. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Complex between the Head-to-Tail Dimer of Calicheamicin Gamma-1-I Oligosaccharide and a DNA Duplex Containing D(ACCT) and D(TCCT) High-Affinity Binding Sites
J.Am.Chem.Soc., 120, 1998
|
|
