1VH4
 
 | |
1VHO
 
 | |
1VHY
 
 | |
1VIY
 
 | | Crystal structure of dephospho-CoA kinase | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
6RPV
 
 | | Extremely stable monomeric variant of human cystatin C with single amino acid substitution | | Descriptor: | Cystatin-C | | Authors: | Zhukov, I, Rodziewicz-Motowidlo, S, Maszota-Zieleniak, M, Jurczak, P, Kozak, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
6G2Z
 
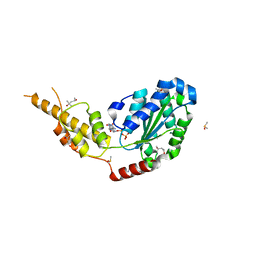 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G4E
 
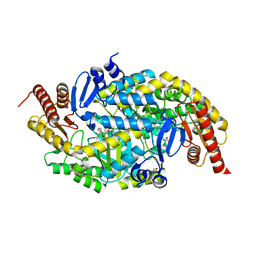 | |
2WTN
 
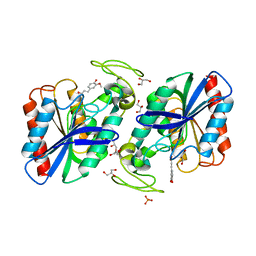 | | Ferulic Acid bound to Est1E from Butyrivibrio proteoclasticus | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, EST1E, GLYCEROL, ... | | Authors: | Goldstone, D.C, Arcus, V.L. | | Deposit date: | 2009-09-17 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
6GDU
 
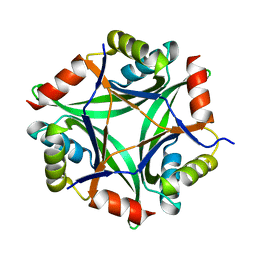 | | Structure of CutA from Synechococcus elongatus PCC7942 | | Descriptor: | Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
6GIR
 
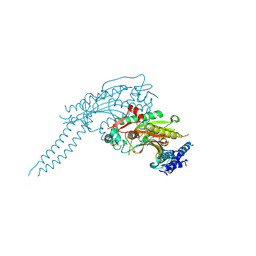 | | Arabidopsis thaliana cytosolic seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Kekez, M, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Arabidopsis seryl-tRNA synthetase: the first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase.
FEBS J., 286, 2019
|
|
6GUJ
 
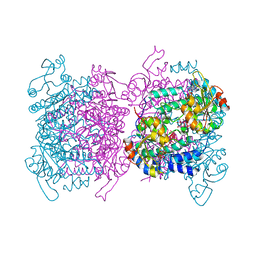 | | Molybdenum storage protein with two occupied ATP binding sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MOLYBDATE ION, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6GG1
 
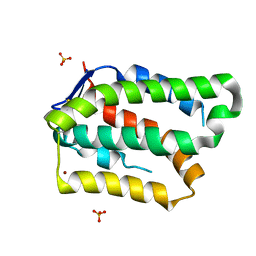 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
5ZNY
 
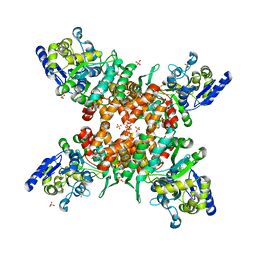 | | Structure of mDR3_DD-C363G with MBP tag | | Descriptor: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5ZNZ
 
 | | Structure of mDR3 DD with MBP tag mutant-I387V | | Descriptor: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | Authors: | Jin, T, Yin, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6DXS
 
 | | Crystal structure of the LigJ hydratase E284Q mutant substrate complex with (3Z)-2-keto-4-carboxy-3-hexenedioate | | Descriptor: | (2Z)-4-oxobut-2-ene-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Hogancamp, T.N. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
4XNK
 
 | | X-ray structure of AlgE1 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alginate production protein AlgE, ... | | Authors: | Ma, P, Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6A6K
 
 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
6A83
 
 | |
5TM0
 
 | | Solution NMR structures of two alternative conformations of E. coli tryptophan repressor in dynamic equilibrium | | Descriptor: | Trp operon repressor | | Authors: | Harish, B, Swapna, G.V.T, Kornhaber, G.J, Montelione, G.T, Carey, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiple helical conformations of the helix-turn-helix region revealed by NOE-restrained MD simulations of tryptophan aporepressor, TrpR.
Proteins, 85, 2017
|
|
6Q0N
 
 | | Structure of the Erbin PDB domain in complex with a high-affinity peptide | | Descriptor: | Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6AEQ
 
 | |
6EGC
 
 | |
1OW5
 
 | |
1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
6AH6
 
 | | M500V mutant of Coronin coiled coil domain | | Descriptor: | Coronin-like protein | | Authors: | Ansari, A, Karade, S.S, Pratap, J.V. | | Deposit date: | 2018-08-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
