1ILY
 
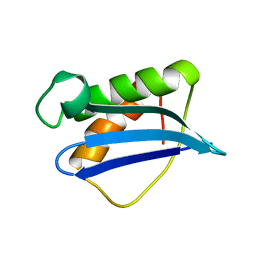 | | Solution Structure of Ribosomal Protein L18 of Thermus thermophilus | | Descriptor: | RIBOSOMAL PROTEIN L18 | | Authors: | Woestenenk, E.A, Gongadze, G.M, Shcherbakov, D.V, Rak, A.V, Garber, M.B, Hard, T, Berglund, H. | | Deposit date: | 2001-05-09 | | Release date: | 2002-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold.
Biochem.J., 363, 2002
|
|
1ILZ
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 6.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1IM0
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT PH 8.3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHSOPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1IM1
 
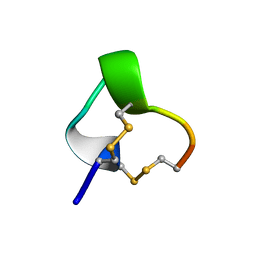 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
1IM2
 
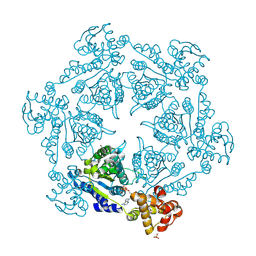 | | HslU, Haemophilus Influenzae, Selenomethionine Variant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2001-05-09 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IM3
 
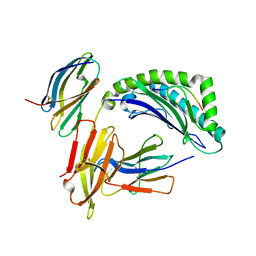 | | Crystal Structure of the human cytomegalovirus protein US2 bound to the MHC class I molecule HLA-A2/tax | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, Human T-cell lymphotropic virus type 1 Tax peptide, ... | | Authors: | Gewurz, B.E, Gaudet, R, Tortorella, D, Wang, E.W, Ploegh, H.L, Wiley, D.C. | | Deposit date: | 2001-05-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IM4
 
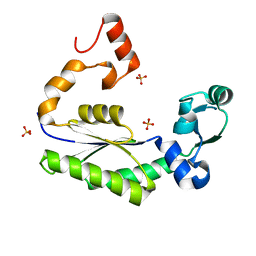 | |
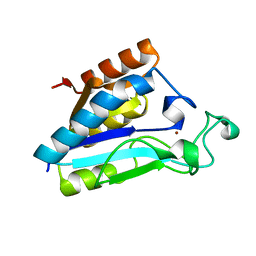
1IM5
 
 | |
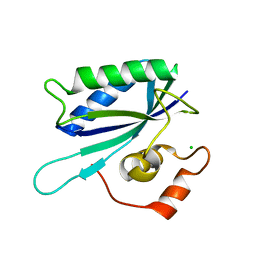
1IM6
 
 | |
1IM7
 
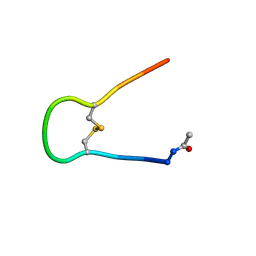 | | Solution structure of synthetic cyclic peptide mimicking the loop of HIV-1 gp41 glycoprotein envelope | | Descriptor: | GP41-PARENT PEPTIDE ACE-ILE-TRP-GLY-CYS-SER-GLY-LYS-LEU-ILE-CYS-THR-THR-ALA | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-10 | | Release date: | 2002-10-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1IM8
 
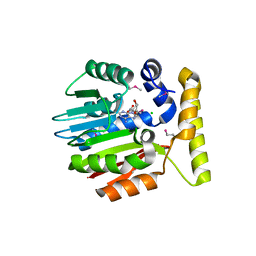 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
1IM9
 
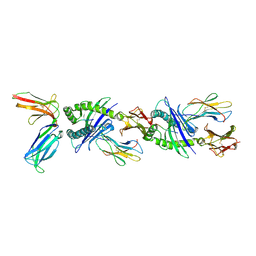 | | Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1 bound to its MHC ligand HLA-Cw4 | | Descriptor: | Beta-2 microglobulin, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, CW-4 CW*0401 ALPHA CHAIN, ... | | Authors: | Fan, Q.R, Long, E.O, Wiley, D.C. | | Deposit date: | 2001-05-10 | | Release date: | 2001-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex.
Nat.Immunol., 2, 2001
|
|
1IMA
 
 | |
1IMB
 
 | |
1IMC
 
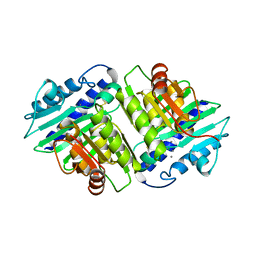 | |
1IMD
 
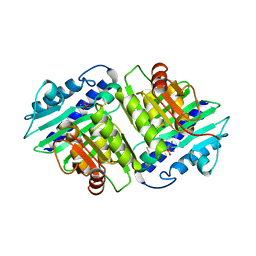 | |
1IME
 
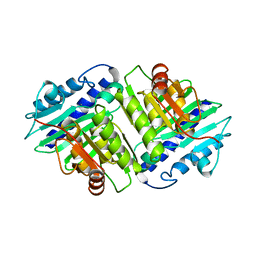 | |
1IMF
 
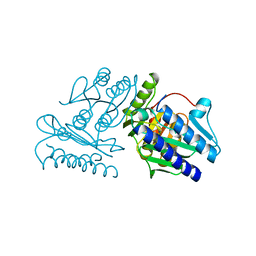 | |
1IMH
 
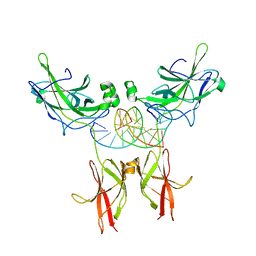 | | TonEBP/DNA COMPLEX | | Descriptor: | 5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*TP*TP*CP*CP*AP*GP*C)-3', 5'-D(*TP*TP*GP*CP*TP*GP*GP*AP*AP*AP*AP*AP*TP*AP*G)-3', NUCLEAR FACTOR OF ACTIVATED T CELLS 5 | | Authors: | Stroud, J.C, Lopez-Rodriguez, C, Rao, A, Chen, L. | | Deposit date: | 2001-05-10 | | Release date: | 2002-02-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of a TonEBP-DNA complex reveals DNA encircled by a transcription factor.
Nat.Struct.Biol., 9, 2002
|
|
1IMI
 
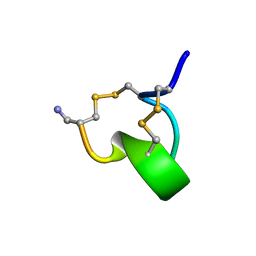 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
1IMJ
 
 | |
1IML
 
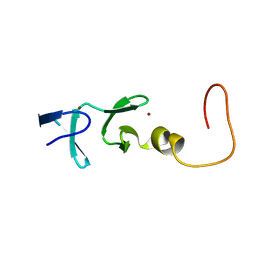 | | CYSTEINE RICH INTESTINAL PROTEIN, NMR, 48 STRUCTURES | | Descriptor: | CYSTEINE RICH INTESTINAL PROTEIN, ZINC ION | | Authors: | Perez-Alvarado, G.C, Kosa, J.L, Louis, H.A, Beckerle, M.C, Winge, D.R, Summers, M.F. | | Deposit date: | 1995-12-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the cysteine-rich intestinal protein, CRIP.
J.Mol.Biol., 257, 1996
|
|
1IMO
 
 | |
1IMP
 
 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, 21 STRUCTURES | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.-Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
1IMQ
 
 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
