3F92
 
 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington Interacting Protein 2) M172A mutant crystallized at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Wilson, R.C, Hughes, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-11-13 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
8WZX
 
 | | Cryo-EM structure of the hamster prion 23-144 fibril at pH 3.7 | | Descriptor: | Major prion protein | | Authors: | Lee, C.-H, Saw, J.-E, Chen, E, Wang, C.-H, Chen, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The Positively Charged Cluster in the N-terminal Disordered Region may Affect Prion Protein Misfolding: Cryo-EM Structure of Hamster PrP(23-144) Fibrils.
J.Mol.Biol., 436, 2024
|
|
6W2H
 
 | | Crystal Structure of the Internal UBA Domain of HHR23A | | Descriptor: | SULFATE ION, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Residual Structure in the Denatured State of the Fast-Folding UBA(1) Domain from the Human DNA Excision Repair Protein HHR23A.
Biochemistry, 61, 2022
|
|
1FDN
 
 | | REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDURICI AT 1.84 ANGSTROMS RESOLUTION | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Duee, E, Fanchon, E, Vicat, J, Sieker, L.C, Meyer, J, Moulis, J-M. | | Deposit date: | 1994-03-31 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution.
J.Mol.Biol., 243, 1994
|
|
3C86
 
 | | OpdA from agrobacterium radiobacter with bound product diethyl thiophosphate from crystal soaking with tetraethyl dithiopyrophosphate- 1.8 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2008-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
2X60
 
 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANNOSE-1-PHOSPHATE GUANYLYLTRANSFERASE | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
4KGB
 
 | |
7KMD
 
 | |
5X13
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5MJ1
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
1CH7
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN H97F MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
2M65
 
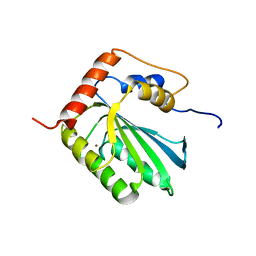 | | NMR structure of human restriction factor APOBEC3A | | Descriptor: | Probable DNA dC->dU-editing enzyme APOBEC-3A, ZINC ION | | Authors: | Byeon, I.L, Byeon, C, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity.
Nat Commun, 4, 2013
|
|
1CP0
 
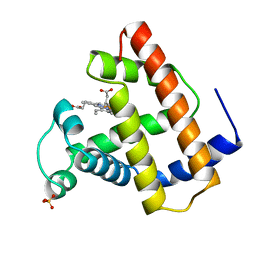 | | RECOMBINANT SPERM WHALE MYOGLOBIN L104N MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
1CPW
 
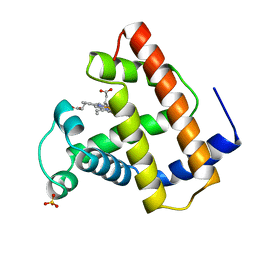 | | RECOMBINANT SPERM WHALE MYOGLOBIN L104W MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
3FAH
 
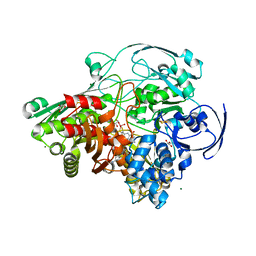 | | Glycerol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Aldehyde oxidoreductase, CHLORIDE ION, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
1CQZ
 
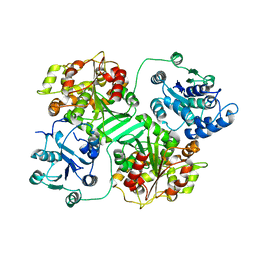 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE. | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3FC4
 
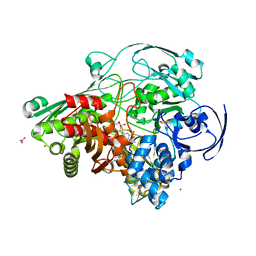 | | Ethylene glycol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 1,2-ETHANEDIOL, Aldehyde oxidoreductase, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
4K28
 
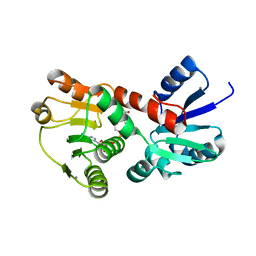 | | 2.15 Angstrom resolution crystal structure of a shikimate dehydrogenase family protein from Pseudomonas putida KT2440 in complex with NAD+ | | Descriptor: | MANGANESE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Garcia, C, Peek, J, Petit, P, Christendat, D. | | Deposit date: | 2013-04-08 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the function of RifI2: structural and biochemical investigation of a new shikimate dehydrogenase family protein.
Biochim.Biophys.Acta, 1834, 2013
|
|
2LZV
 
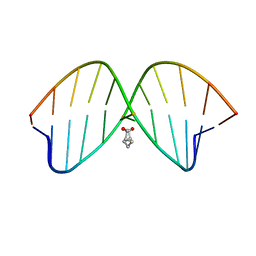 | | DNA duplex containing mispair-aligned O4U-heptylene-O4U interstrand cross-link | | Descriptor: | DNA (5'-D(*CP*GP*AP*AP*AP*UP*TP*TP*TP*CP*G)-3'), HEPTANE | | Authors: | Denisov, A.Y, McManus, F.P, Noronha, A.M, Wilds, C.J. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of interstrand cross-link repair by O 6 -alkylguanine DNA alkyltransferase.
Org.Biomol.Chem., 15, 2017
|
|
2YUU
 
 | | Solution structure of the first Phorbol esters/diacylglycerol binding domain of human Protein kinase C, delta | | Descriptor: | Protein kinase C delta type, ZINC ION | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Saito, K, Sasagawa, A, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first Phorbol esters/diacylglycerol binding domain of human Protein kinase C, delta
To be Published
|
|
5KVG
 
 | | Zika specific antibody, ZV-67, bound to ZIKA envelope DIII | | Descriptor: | CHLORIDE ION, ZIKA Envelope DIII, ZV-67 Antibody Fab Heavy Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
2PKU
 
 | | Solution structure of PICK1 PDZ in complex with the carboxyl tail peptide of GluR2 | | Descriptor: | PRKCA-binding protein, peptide (GLU)(SER)(VAL)(LYS)(ILE) | | Authors: | Pan, L, Wu, H, Shen, C, Shi, Y, Xia, J, Zhang, M. | | Deposit date: | 2007-04-18 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes
Embo J., 26, 2007
|
|
5OQK
 
 | | Solution NMR structure of truncated, human Hv1/VSOP (Voltage-gated proton channel) | | Descriptor: | Voltage-gated hydrogen channel 1 | | Authors: | Bayrhuber, M, Maslennikov, I, Kwiatowski, W, Sobol, A, Wierschem, C, Eichmann, C, Riek, R. | | Deposit date: | 2017-08-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure and Functional Behavior of the Human Proton Channel.
Biochemistry, 58, 2019
|
|
6W9D
 
 | |
2K19
 
 | | NMR solution structure of PisI | | Descriptor: | Putative piscicolin 126 immunity protein | | Authors: | Martin-Visscher, L.A, Sprules, T, Gursky, L.J, Vederas, J.C. | | Deposit date: | 2008-02-25 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of PisI, a group B immunity protein that provides protection against the type IIa bacteriocin piscicolin 126, PisA.
Biochemistry, 47, 2008
|
|
