5CET
 
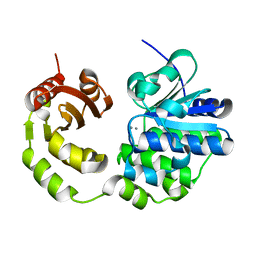 | | Crystal structure of Rv2837c | | Descriptor: | Bifunctional oligoribonuclease and PAP phosphatase NrnA, MANGANESE (II) ION | | Authors: | Wang, F, He, Q, Zhu, D, Liu, S, Gu, L. | | Deposit date: | 2015-07-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Insight into the Mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP Phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
6KSD
 
 | |
2XIX
 
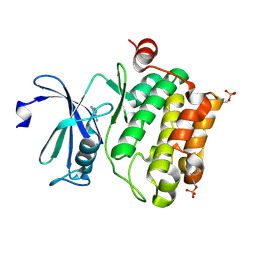 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ2
 
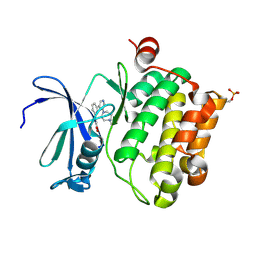 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XIY
 
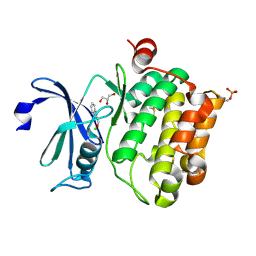 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZUF
 
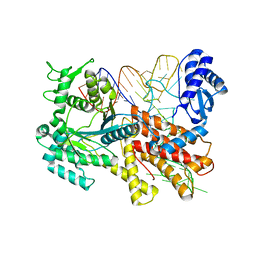 | | Crystal structure of Pyrococcus horikoshii arginyl-tRNA synthetase complexed with tRNA(Arg) | | Descriptor: | Arginyl-tRNA synthetase, tRNA-Arg | | Authors: | Konno, M, Sumida, T, Uchikawa, E, Mori, Y, Yanagisawa, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP)
Febs J., 276, 2009
|
|
2FSG
 
 | |
5XSP
 
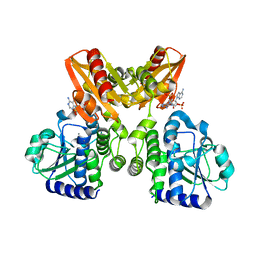 | | The catalytic domain of GdpP with 5'-pApA | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
4YXW
 
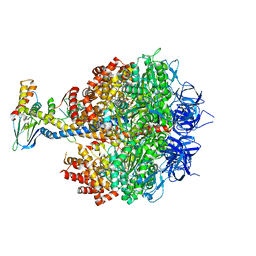 | | Bovine heart mitochondrial F1-ATPase inhibited by AMP-PNP and ADP in the presence of thiophosphate. | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3E7X
 
 | | Crystal structure of DLTA: implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1 | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
3E7W
 
 | | Crystal structure of DLTA: Implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1, PHOSPHATE ION | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
4OHY
 
 | | C. Elegans Clp1 bound to ssRNA dinucleotide GC, AMP-PNP, and Mg2+(inhibited substrate bound state) | | Descriptor: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
4OHV
 
 | | C. Elegans Clp1 bound to AMP-PNP, and Mg2+ | | Descriptor: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
2GCQ
 
 | |
4R0M
 
 | | Structure of McyG A-PCP complexed with phenylalanyl-adenylate | | Descriptor: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], McyG protein | | Authors: | Tan, X.F, Dai, Y.N, Zhou, K, Jiang, Y.L, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the adenylation-peptidyl carrier protein didomain of the Microcystis aeruginosa microcystin synthetase McyG.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1MBZ
 
 | | BETA-LACTAM SYNTHETASE WITH TRAPPED INTERMEDIATE | | Descriptor: | ARGININE-N-METHYLCARBONYL PHOSPHORIC ACID 5'-ADENOSINE ESTER, BETA-LACTAM SYNTHETASE, GLYCEROL, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2002-08-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3KYC
 
 | | Human SUMO E1 complex with a SUMO1-AMP mimic | | Descriptor: | 5'-deoxy-5'-(sulfamoylamino)adenosine, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
2XMI
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, complexed with citrate | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, CITRATE ANION, ... | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Myelin 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase: Active-Site Ligand Binding and Molecular Conformation.
Plos One, 7, 2012
|
|
3KYD
 
 | | Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, SUMO-activating enzyme subunit 1, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
2Y3X
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, complexed with sulfate | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, GLYCEROL, ... | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2011-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Myelin 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase: Active-Site Ligand Binding and Molecular Conformation.
Plos One, 7, 2012
|
|
2Y1P
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, complexed with citrate | | Descriptor: | 2', 3'-CYCLIC NUCLEOTIDE 3'-PHOSPHODIESTERASE, CITRIC ACID, ... | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Myelin 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase: Active-Site Ligand Binding and Molecular Conformation.
Plos One, 7, 2012
|
|
3CJ1
 
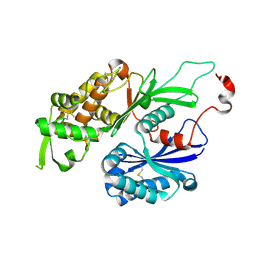 | | Structure of Rattus norvegicus NTPDase2 | | Descriptor: | Ectonucleoside triphosphate diphosphohydrolase 2 | | Authors: | Zebisch, M, Strater, N. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6DZG
 
 | |
1AKE
 
 | |
2ARU
 
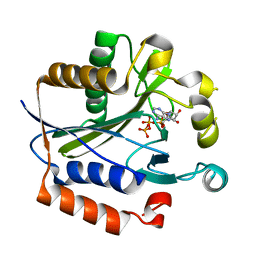 | | Crystal structure of lipoate-protein ligase A bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
