2GIL
 
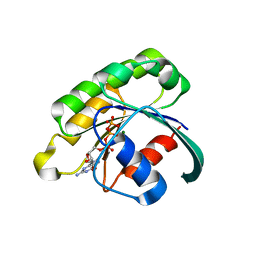 | | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6A | | Authors: | Bergbrede, T, Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution
J.STRUCT.BIOL., 152, 2005
|
|
6KWH
 
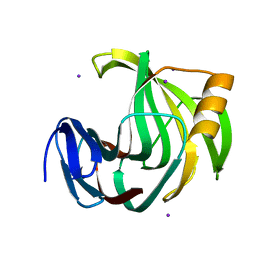 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism
To Be Published
|
|
7DKX
 
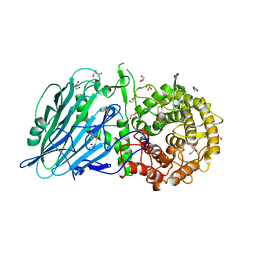 | |
7DKU
 
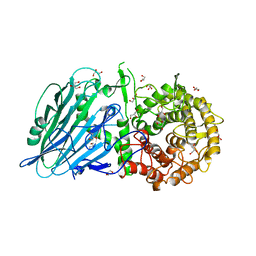 | |
7DKW
 
 | |
2G8Q
 
 | |
2GD6
 
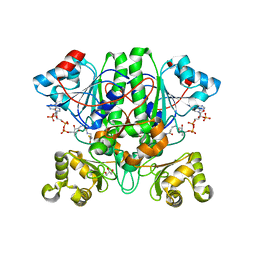 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
7DKV
 
 | |
2GDI
 
 | |
5A7U
 
 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
7DKY
 
 | |
2GET
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-I (LT) | | Descriptor: | GLYCEROL, Pantothenate kinase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-4-{[3-({2-[(2-HYDROXYETHYL)DITHIO]ETHYL}AMINO)-3-OXOPROPYL]AMINO}-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5ABG
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-1-[3-(4-fluorophenyl)propyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2GFK
 
 | | Crystal structure of the zinc-beta-lactamase l1 from stenotrophomonas maltophilia (inhibitor 2) | | Descriptor: | 2,5-DIPHENYLFURAN-3,4-DICARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, Metallo-beta-lactamase L1, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
2GEV
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-II (LT) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Pantothenate kinase, ... | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5AI7
 
 | | ParM doublet model | | Descriptor: | PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles
Nature, 523, 2015
|
|
5AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'RED' SUBSTATE, AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
2GGP
 
 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
5A9K
 
 | | Structural basis for DNA strand separation by a hexameric replicative helicase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, REPLICATION PROTEIN E1 | | Authors: | Chaban, Y, Stead, J.A, Ryzhenkova, K, Whelan, F, Lamber, K, Antson, F, Sanders, C.M, Orlova, E.V. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Structural Basis for DNA Strand Separation by a Hexameric Replicative Helicase.
Nucleic Acids Res., 43, 2015
|
|
4ZVD
 
 | |
2G45
 
 | | Co-crystal structure of znf ubp domain from the deubiquitinating enzyme isopeptidase T (isot) in complex with ubiquitin | | Descriptor: | CHLORIDE ION, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
5ACA
 
 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AJI
 
 | | MscS D67R1 high resolution | | Descriptor: | DECANE, HEXANE, N-OCTANE, ... | | Authors: | Naismith, J.H, Pliotas, C. | | Deposit date: | 2015-02-24 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Role of Lipids in Mechanosensation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4ZWU
 
 | | Crystal structure of organophosphate anhydrolase/prolidase mutant Y212F, V342L, I215Y | | Descriptor: | BARIUM ION, GLYCEROL, GLYCOLIC ACID, ... | | Authors: | Daczkowski, C.M, Pegan, S.D, Harvey, S.P. | | Deposit date: | 2015-05-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering the Organophosphorus Acid Anhydrolase Enzyme for Increased Catalytic Efficiency and Broadened Stereospecificity on Russian VX.
Biochemistry, 54, 2015
|
|
4ZXA
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Cd2+ and 4-hydroxybenzonitrile | | Descriptor: | 4-hydroxybenzonitrile, CADMIUM ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
