6U1Q
 
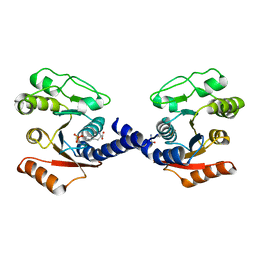 | | Crystal Structure of VpsO (VC0937) Kinase domain | | Descriptor: | O-PHOSPHOTYROSINE, VpsO | | Authors: | Tripathi, S.M, Schwechheimer, C, Herbert, K, Porcella, M.E, Brown, E.R, Yildiz, F.H, Rubin, S.M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae.
Plos Pathog., 16, 2020
|
|
3VE5
 
 | |
4ZUN
 
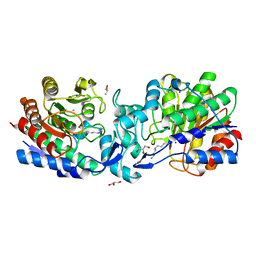 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a thiol inhibitor | | Descriptor: | 5-[(3-aminopropyl)amino]pentane-1-thiol, Acetylpolyamine aminohydrolase, GLYCEROL, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
1E58
 
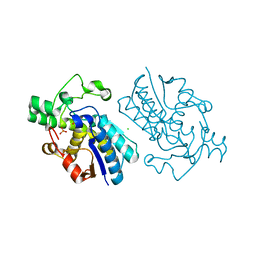 | | E.coli cofactor-dependent phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCERATE MUTASE, SULFATE ION | | Authors: | Bond, C.S, Hunter, W.N. | | Deposit date: | 2000-07-19 | | Release date: | 2001-03-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Structure of the Phosphohistidine-Activated Form of Escherichia Coli Cofactor-Dependent Phosphoglycerate Mutase.
J.Biol.Chem., 276, 2001
|
|
3L4R
 
 | | Crystal structure of the dog lipocalin allergen Can f 2 and implications for cross-reactivity to the cat allergen Fel d 4 | | Descriptor: | GLYCEROL, Minor allergen Can f 2 | | Authors: | Madhurantakam, C, Nilsson, O.B, Gronlund, H, Achour, A. | | Deposit date: | 2009-12-21 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Dog Lipocalin Allergen Can f 2: Implications for Cross-reactivity to the Cat Allergen Fel d 4
J.Mol.Biol., 401, 2010
|
|
6NK2
 
 | |
8CSR
 
 | | Human mitochondrial small subunit assembly intermediate (State C) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
1HXI
 
 | | AN UNEXPECTED EXTENDED CONFORMATION FOR THE THIRD TPR MOTIF OF THE PEROXIN PEX5 FROM TRYPANOSOMA BRUCEI | | Descriptor: | MAGNESIUM ION, PEROXISOME TARGETING SIGNAL 1 RECEPTOR PEX5 | | Authors: | Kumar, A, Roach, C, Hirsh, I.S, Turley, S, deWalque, S, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2001-01-15 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei.
J.Mol.Biol., 307, 2001
|
|
8CSU
 
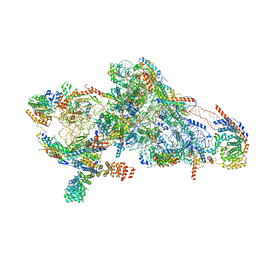 | | Human mitochondrial small subunit assembly intermediate (State C*) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
1ZCL
 
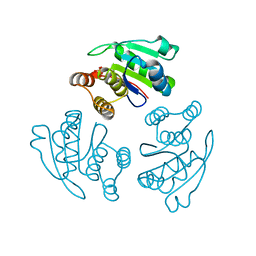 | | prl-1 c104s mutant in complex with sulfate | | Descriptor: | SULFATE ION, protein tyrosine phosphatase 4a1 | | Authors: | Sun, J.P, Wang, W.Q, Yang, H, Liu, S, Liang, F, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion.
Biochemistry, 44, 2005
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
7TI1
 
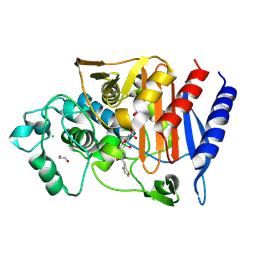 | | Structure of AmpC bound to RPX-7063 at 2.0A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Abendroth, J, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
3FIS
 
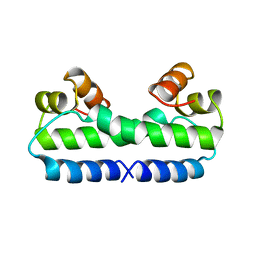 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
6GVB
 
 | | Crystal structure of Cutibacterium acnes exo-beta-1,4-mannosidase | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, exo-beta-1,4-mannosidase | | Authors: | Reichenbach, T, Divne, C. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical characterization of the Cutibacterium acnes exo-beta-1,4-mannosidase that targets the N-glycan core of host glycoproteins.
Plos One, 13, 2018
|
|
5M6E
 
 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[3,3-dimethyl-6-(phenylmethyl)-2~{H}-pyrrolo[3,2-c]pyridin-1-yl]-2-[(2~{R},5~{R})-5-methyl-2-[(4-methylpyrazol-1-yl)methyl]piperazin-4-ium-1-yl]ethanone, DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase XIAP, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
6HY3
 
 | | Three-dimensional structure of AgaC from Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, Beta-agarase C, GLYCEROL, ... | | Authors: | Naretto, A, Fanuel, M, Ropartz, D, Rogniaux, H, Larocque, R, Czjzek, M, Tellier, C, Michel, G. | | Deposit date: | 2018-10-19 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The agar-specific hydrolaseZgAgaC from the marine bacteriumZobellia galactanivoransdefines a new GH16 protein subfamily.
J.Biol.Chem., 294, 2019
|
|
8R4C
 
 | |
7NEN
 
 | |
1CP2
 
 | | NITROGENASE IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Schlessman, J.L, Woo, D, Joshua-Tor, L, Howard, J.B, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
8EQM
 
 | | Structure of a dimeric photosystem II complex acclimated to far-red light | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Shen, G, Flesher, D.A, Kurashov, V, Golbeck, J.H, Brudvig, G.W, Amin, M, Bryant, D.A. | | Deposit date: | 2022-10-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a dimeric photosystem II complex from a cyanobacterium acclimated to far-red light.
J.Biol.Chem., 299, 2022
|
|
8R4B
 
 | |
5C22
 
 | | Crystal structure of Zn-bound HlyD from E. coli | | Descriptor: | Chromosomal hemolysin D, ZINC ION | | Authors: | Ha, N.C, Kim, J.S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
7EB0
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
5LFX
 
 | |
3ML0
 
 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in tetragonal form | | Descriptor: | CALCIUM ION, Penicillin G acylase, alpha subunit, ... | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
