1B4R
 
 | | PKD DOMAIN 1 FROM HUMAN POLYCYSTEIN-1 | | Descriptor: | PROTEIN (PKD1_HUMAN) | | Authors: | Bycroft, M. | | Deposit date: | 1998-12-28 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease.
EMBO J., 18, 1999
|
|
1B4S
 
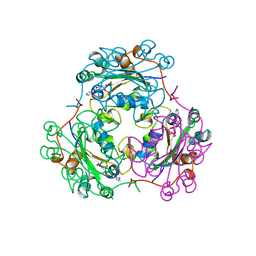 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE H122G MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE, ... | | Authors: | Meyer, P, Janin, J. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase.
Biochemistry, 38, 1999
|
|
1FAK
 
 | | HUMAN TISSUE FACTOR COMPLEXED WITH COAGULATION FACTOR VIIA INHIBITED WITH A BPTI-MUTANT | | Descriptor: | CALCIUM ION, PROTEIN (5L15), PROTEIN (BLOOD COAGULATION FACTOR VIIA), ... | | Authors: | Zhang, E, St Charles, R, Tulinsky, A. | | Deposit date: | 1998-12-28 | | Release date: | 1999-12-03 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of extracellular tissue factor complexed with factor VIIa inhibited with a BPTI mutant.
J.Mol.Biol., 285, 1999
|
|
1FZF
 
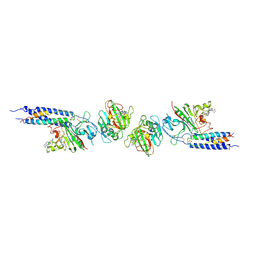 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
2AIY
 
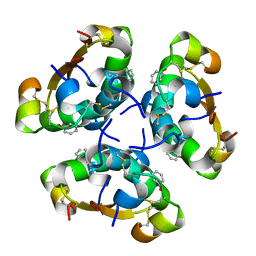 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 20 STRUCTURES | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-28 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
438D
 
 | |
1JOY
 
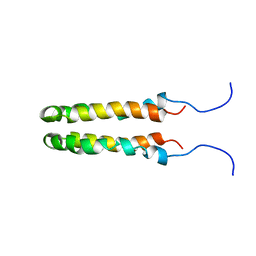 | | SOLUTION STRUCTURE OF THE HOMODIMERIC DOMAIN OF ENVZ FROM ESCHERICHIA COLI BY MULTI-DIMENSIONAL NMR. | | Descriptor: | PROTEIN (ENVZ_ECOLI) | | Authors: | Tomomori, C, Tanaka, T, Dutta, R, Park, H, Saha, S.K, Zhu, Y, Ishima, R, Liu, D, Tong, K.I, Kurokawa, H, Qian, H, Inouye, M, Ikura, M. | | Deposit date: | 1998-12-28 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ.
Nat.Struct.Biol., 6, 1999
|
|
1B4U
 
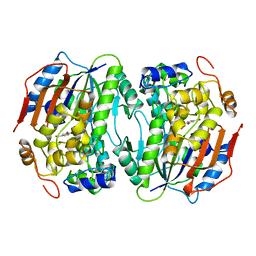 | | PROTOCATECHUATE 4,5-DIOXYGENASE (LIGAB) IN COMPLEX WITH PROTOCATECHUATE (PCA) | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 4,5-DIOXYGENASE | | Authors: | Sugimoto, K, Senda, T, Mitsui, Y. | | Deposit date: | 1998-12-29 | | Release date: | 1999-08-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
Structure Fold.Des., 7, 1999
|
|
2SRC
 
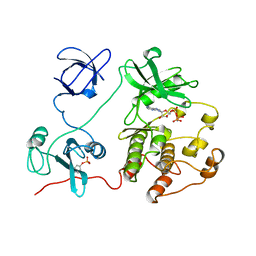 | | CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TYROSINE-PROTEIN KINASE SRC | | Authors: | Xu, W, Doshi, A, Lei, M, Eck, M.J, Harrison, S.C. | | Deposit date: | 1998-12-29 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of c-Src reveal features of its autoinhibitory mechanism.
Mol.Cell, 3, 1999
|
|
5AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'RED' SUBSTATE, AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
4AIY
 
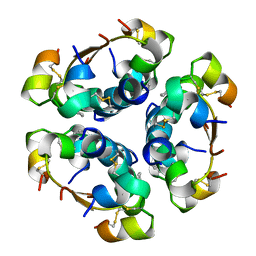 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'GREEN' SUBSTATE, AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
5EAA
 
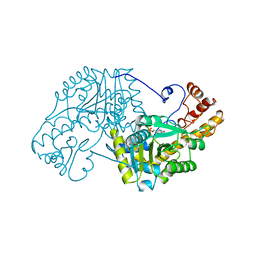 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-29 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
3AIY
 
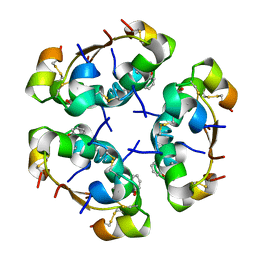 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
1B4X
 
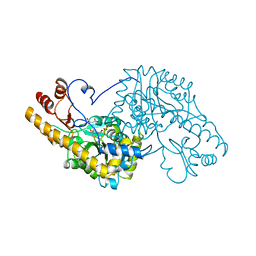 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-30 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
1B4W
 
 | |
1B4V
 
 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1998-12-30 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
1B4Y
 
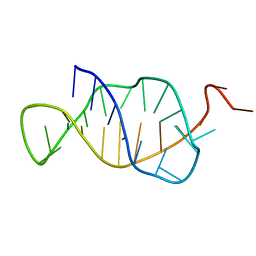 | | STRUCTURE AND MECHANISM OF FORMATION OF THE H-Y5 ISOMER OF AN INTRAMOLECULAR DNA TRIPLE HELIX. | | Descriptor: | DNA (H-Y5 TRIPLE HELIX) | | Authors: | Van Dongen, M.J.P, Doreleijers, J.F, Van Der Marel, G.A, Van Boom, J.H, Hilbers, C.W, Wijmenga, S.S. | | Deposit date: | 1998-12-30 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix.
Nat.Struct.Biol., 6, 1999
|
|
1B7U
 
 | |
1FZG
 
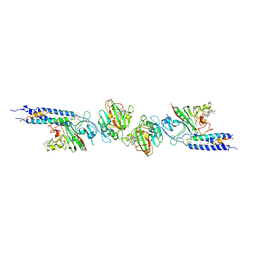 | | CRYSTAL STRUCTURE OF FRAGMENT D FROM HUMAN FIBRINOGEN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1999-01-01 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
2BTM
 
 | |
1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
1B4Z
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KDK | | Descriptor: | ACETATE ION, PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), PROTEIN (PEPTIDE LYS-ASP-LYS), ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and calorimetric analysis of peptide binding to OppA protein.
J.Mol.Biol., 291, 1999
|
|
2HGS
 
 | | HUMAN GLUTATHIONE SYNTHETASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, MAGNESIUM ION, ... | | Authors: | Polekhina, G, Board, P, Rossjohn, J, Parker, M.W. | | Deposit date: | 1999-01-04 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of glutathione synthetase deficiency and a rare gene permutation event.
EMBO J., 18, 1999
|
|
3MCT
 
 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3CYT AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 3-METHYLCYTOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Gershon, P.D, Hodel, A.E, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1B42
 
 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M1ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-1-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
