4JK7
 
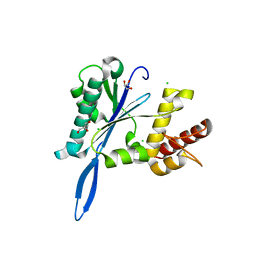 | | Open and closed forms of wild-type human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKD
 
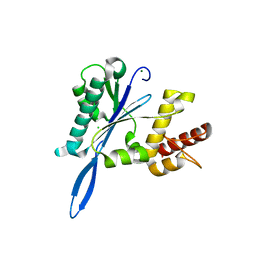 | | Open and closed forms of I1790Y human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKC
 
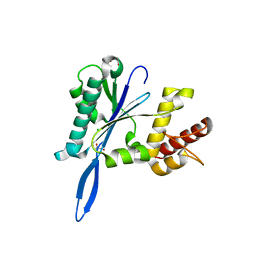 | | Open and closed forms of T1800E human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
161D
 
 | |
4JK8
 
 | | Open and closed forms of R1865A human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JK9
 
 | | Open and closed forms of wild-type human PRP8 RNase H-like domain with bound Co ion | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKH
 
 | | Open and closed forms of D1781E human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKB
 
 | | Open and closed forms of V1788D human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKA
 
 | | Open and closed forms of R1865A human PRP8 RNase H-like domain with bound Co ion | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKF
 
 | | Open and closed forms of T1791P+R1865A human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKE
 
 | | Open and closed forms of T1789P human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
9KZP
 
 | | Z-form DNA-RNA hybrid in dynamic simulation | | Descriptor: | DNA (5'-D(*(5CM)P*GP*(5CM)P*AP*(5CM)P*GP*(5CM)P*G)-3'), RNA (5'-R(*CP*GP*CP*GP*UP*GP*CP*G)-3') | | Authors: | Wang, S.Y, Xu, Y. | | Deposit date: | 2024-12-11 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Z-form DNA-RNA hybrid blocks DNA replication.
Nucleic Acids Res., 53, 2025
|
|
1FMN
 
 | | SOLUTION STRUCTURE OF FMN-RNA APTAMER COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*CP*GP*UP*GP*UP*AP*GP*GP *AP*UP*AP*UP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*AP*AP*GP *GP*AP*CP*AP*CP*GP*CP*C)-3') | | Authors: | Fan, P, Suri, A.K, Fiala, R, Live, D, Patel, D.J. | | Deposit date: | 1995-12-04 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the FMN-RNA aptamer complex.
J.Mol.Biol., 258, 1996
|
|
2AG6
 
 | | Crystal structure of p-bromo-l-phenylalanine-tRNA sythetase in complex with p-bromo-l-phenylalanine | | Descriptor: | 4-BROMO-L-PHENYLALANINE, Tyrosyl-tRNA synthetase | | Authors: | Turner, J.M, Graziano, J, Spraggon, G, Schultz, P.G. | | Deposit date: | 2005-07-26 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of an aminoacyl-tRNA synthetase active site
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5IPP
 
 | |
1ZH6
 
 | | Crystal Structure of p-acetylphenylalanine-tRNA synthetase in complex with p-acetylphenylalanine | | Descriptor: | 4-ACETYL-L-PHENYLALANINE, BETA-MERCAPTOETHANOL, Tyrosyl-tRNA synthetase | | Authors: | Turner, J.M, Graziano, J, Spraggon, G, Schultz, P.G. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a p-acetylphenylalanyl aminoacyl-tRNA synthetase.
J.Am.Chem.Soc., 127, 2005
|
|
5IZO
 
 | |
5J21
 
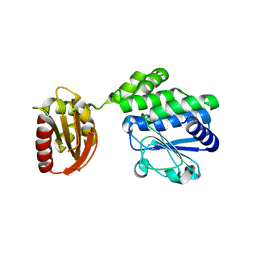 | |
5IUF
 
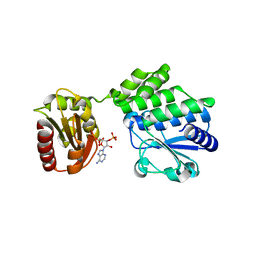 | |
2W4S
 
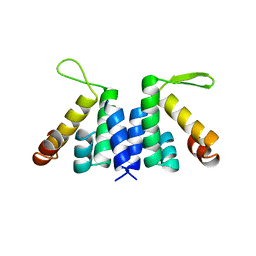 | | novel RNA-binding domain in Cryptosporidium parvum at 2.5 angstrom resolution | | Descriptor: | Ankyrin-repeat protein | | Authors: | Varrot, A, Mackereth, C, Mourao, A, Sattler, M, Cusack, S. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and RNA Recognition by the Snrna and Snorna Transport Factor Phax.
RNA, 16, 2010
|
|
2MFF
 
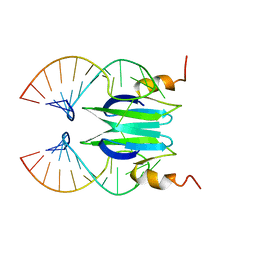 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
7YPW
 
 | | Lloviu cuevavirus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.0356 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
1GTR
 
 | |
2MFE
 
 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL2)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL2(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
7D7U
 
 | | Crystal structure of Ago2 MID domain in complex with 8-Br-adenosin-5'-monophosphate | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-10-06 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
