3HT6
 
 | | 2-methylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | Lysozyme, PHOSPHATE ION, o-cresol | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTD
 
 | | (Z)-Thiophene-2-carboxaldoxime in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (NZ)-N-(thiophen-2-ylmethylidene)hydroxylamine, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
1N7T
 
 | | ERBIN PDZ domain bound to a phage-derived peptide | | Descriptor: | 99-mer peptide of densin-180-like protein, phage-derived peptide | | Authors: | Skelton, N.J, Koehler, M.F.T, Zobel, K, Wong, W.L, Yeh, S, Pisabarro, M.T, Yin, J.P, Lasky, L.A, Sidhu, S.S. | | Deposit date: | 2002-11-16 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain.
J.Biol.Chem., 278, 2003
|
|
6DQT
 
 | | Crystal structure of Haemophilus influenzae OppA complex with LGG | | Descriptor: | ACETATE ION, LEU-GLY-GLY, Periplasmic oligopeptide-binding protein, ... | | Authors: | Tanaka, K.J, Pinkett, H.W. | | Deposit date: | 2018-06-11 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligopeptide-binding protein from nontypeableHaemophilus influenzaehas ligand-specific sites to accommodate peptides and heme in the binding pocket.
J. Biol. Chem., 294, 2019
|
|
7K3O
 
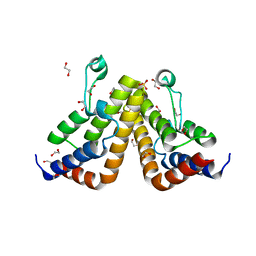 | |
3I7P
 
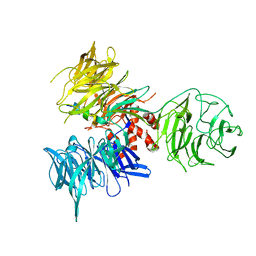 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR40A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 40A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2J3S
 
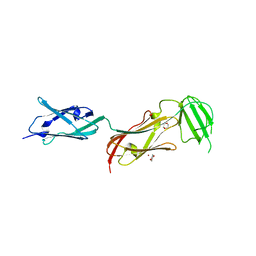 | |
6C9W
 
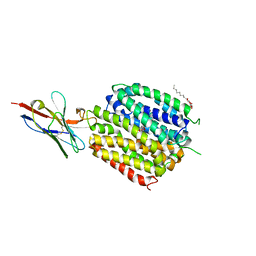 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3I7O
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of IQWD1 | | Descriptor: | DNA damage-binding protein 1, IQ motif and WD repeat-containing protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4Y13
 
 | | SdiA in complex with octanoyl-rac-glycerol | | Descriptor: | (2S)-2,3-dihydroxypropyl octanoate, GLYCEROL, SULFATE ION, ... | | Authors: | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
4E0W
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
4NUX
 
 | | Structure of receptor A | | Descriptor: | Interleukin-17 receptor A | | Authors: | Zhang, B, Han, Y, Deng, J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Structure of the unique SEFIR domain from human interleukin 17 receptor A reveals a composite ligand-binding site containing a conserved alpha-helix for Act1 binding and IL-17 signaling.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3I7N
 
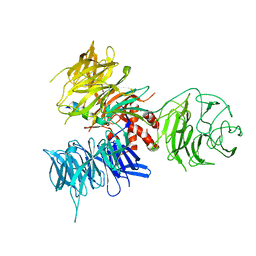 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDTC1 | | Descriptor: | DNA damage-binding protein 1, WD and tetratricopeptide repeats protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4Y17
 
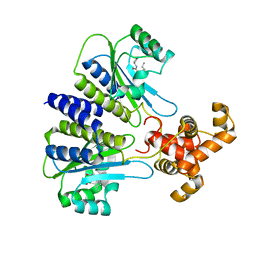 | | SdiA in complex with 3-oxo-C8-homoserine lactone | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, Transcriptional regulator of ftsQAZ gene cluster | | Authors: | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
7D7X
 
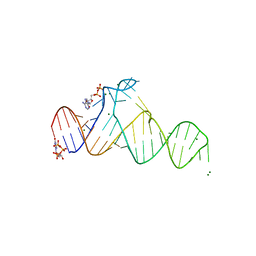 | |
7D7W
 
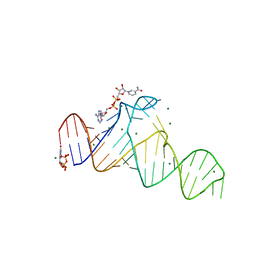 | |
7D82
 
 | |
7D81
 
 | |
4B7G
 
 | | Structure of a bacterial catalase | | Descriptor: | CATALASE, CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
7TWR
 
 | |
7TWP
 
 | |
3I7H
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of HBX | | Descriptor: | DNA damage-binding protein 1, X protein | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7TWF
 
 | |
7TWQ
 
 | |
