8DS3
 
 | | LRRC8A:C conformation 1 (round) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4XJF
 
 | | X-ray structure of Lysozyme B1 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6S2X
 
 | | Crystal structure of the Legionella pneumophila ChiA C-terminal domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ChiA | | Authors: | Garnett, J.A, Shaw, R. | | Deposit date: | 2019-06-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and functional analysis of the Legionella pneumophila chitinase ChiA reveals a novel mechanism of metal-dependent mucin degradation.
Plos Pathog., 16, 2020
|
|
9FGJ
 
 | | Cryo-EM structure of Legionella effector SdeC (PDE-mART domain) | | Descriptor: | SdeC | | Authors: | Weng, T.-H, Misra, M, Chen, W, Safarian, S, Kudryashev, M, Dikic, I. | | Deposit date: | 2024-05-24 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | BAR-like C-terminal domain of Legionella SidE family is crucial for its membrane localization and secretion
To Be Published
|
|
7QBS
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, S-adenosyl-L-methionine, and peptide bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Bernardo-Garcia, N, Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
8BRO
 
 | | Structure of the N-terminal domain of BC2L-C lectin (1-131) in complex with a synthetic beta-fucosylamide | | Descriptor: | 5-[3-(aminomethyl)phenyl]-~{N}-[(2~{S},3~{S},4~{R},5~{S},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]furan-2-carboxamide, Lectin | | Authors: | Varrot, A, Lunstrom, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of New L-Fucosyl and L-Galactosyl Amides as Glycomimetic Ligands of TNF Lectin Domain of BC2L-C from Burkholderia cenocepacia.
Molecules, 28, 2023
|
|
6G1C
 
 | |
7PV8
 
 | |
5V0K
 
 | | RNA duplex with 2-MeImpG analogue bound-3 binding sites | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*C)-3') | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA duplex with 2-MeImpG analogue bound-3 binding sites
To Be Published
|
|
2WFB
 
 | | High resolution structure of the apo form of the orange protein (ORP) from Desulfovibrio gigas | | Descriptor: | ACETATE ION, PHOSPHATE ION, PUTATIVE UNCHARACTERIZED PROTEIN ORP | | Authors: | Najmudin, S, Bonifacio, C, Duarte, A.G, Pereira, A.S, Moura, I, Moura, J.M, Romao, M.J. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Crystal Structure of the Apo Form of the Orange Protein (Apo-Orp) from Desulfovibrio Gigas
To be Published
|
|
6UMA
 
 | |
6RKS
 
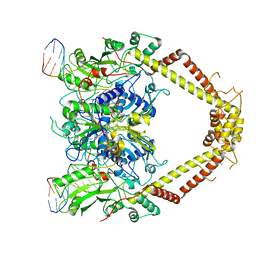 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6UMB
 
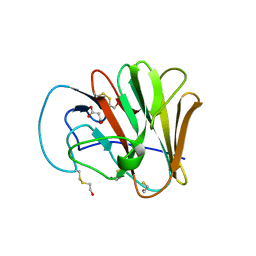 | |
6RPN
 
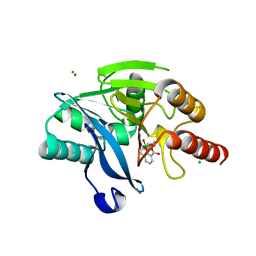 | | Structure of metallo beta lactamase VIM-2 with cyclic boronate APC308. | | Descriptor: | (3~{R})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, (3~{S})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, Beta-lactamase VIM-2, ... | | Authors: | Parkova, A, Lucic, A, Brem, J, McDonough, M.A, Langley, G.W, Schofield, C.J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Broad Spectrum beta-Lactamase Inhibition by a Thioether Substituted Bicyclic Boronate.
Acs Infect Dis., 6, 2020
|
|
8V10
 
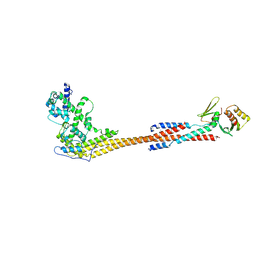 | |
2WJH
 
 | | Structure and function of the FeoB G-domain from Methanococcus jannaschii | | Descriptor: | CITRATE ANION, FERROUS IRON TRANSPORT PROTEIN B HOMOLOG, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Koester, S, Wehner, M, Herrmann, C, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2009-05-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of the Feob G-Domain from Methanococcus Jannaschii
J.Mol.Biol., 392, 2009
|
|
5NIH
 
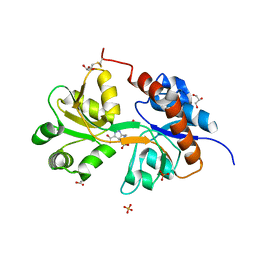 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist LM-12b at 1.3 A resolution. | | Descriptor: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
6G79
 
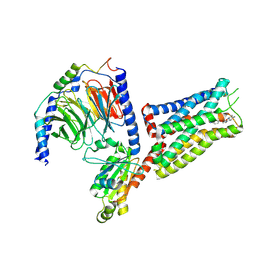 | | Coupling specificity of heterotrimeric Go to the serotonin 5-HT1B receptor | | Descriptor: | 2-[5-[2-[4-(4-cyanophenyl)piperazin-1-yl]-2-oxidanylidene-ethoxy]-1~{H}-indol-3-yl]ethylazanium, 5-hydroxytryptamine receptor 1B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Garcia-Nafria, J, Nehme, R, Edwards, P, Tate, C.G. | | Deposit date: | 2018-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM structure of the serotonin 5-HT1Breceptor coupled to heterotrimeric Go.
Nature, 558, 2018
|
|
2WIY
 
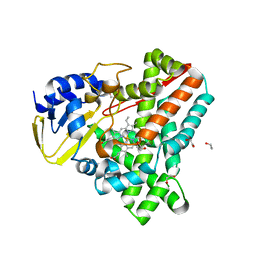 | | Cytochrome P450 XplA heme domain P21212 | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450-LIKE PROTEIN XPLA, IMIDAZOLE, ... | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
4XA3
 
 | | Crystal structure of the coiled-coil surrounding Skip 2 of MYH7 | | Descriptor: | Gp7-MYH7(1361-1425)-Eb1 chimera protein | | Authors: | Taylor, K.C, Buvoli, M, Korkmaz, E.N, Buvoli, A, Zheng, Y, Heinz, N.T, Qiang, C, Leinwand, L.A, Rayment, I. | | Deposit date: | 2014-12-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2WIV
 
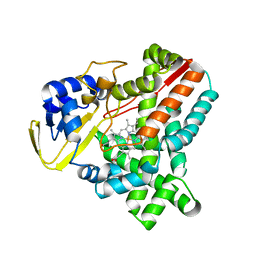 | | Cytochrome-P450 XplA heme domain P21 | | Descriptor: | CYTOCHROME P450-LIKE PROTEIN XPLA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
7OA3
 
 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium hexammine co-crystallized form) | | Descriptor: | Chili RNA Aptamer, DMHBO+, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
6RIV
 
 | | Crystal structure of Alopecurus myosuroides GSTF | | Descriptor: | GLYCEROL, Glutathione transferase, S-Hydroxy-Glutathione, ... | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2019-04-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Comparative structural and functional analysis of phi class glutathione transferases involved in multiple-herbicide resistance of grass weeds and crops.
Plant Physiol Biochem., 149, 2020
|
|
8SR2
 
 | | particulate methane monooxygenase incubated with 4,4,4-trifluorobutanol | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
