4OO4
 
 | |
5RC2
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E11b | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6YP6
 
 | |
5RE1
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 58 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1VL9
 
 | | Atomic resolution (0.97A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Gayathri, D, Poi, M.-J, Tsai, M.-D, Dauter, M, Dauter, Z. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
6JJ1
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Iqbal, N, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-24 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues
To Be Published
|
|
5JEU
 
 | | del-[Ru(phen)2(dppz)]2+ bound to d(TCGGCGCCGA) with Ba2+ | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Delta chirality ruthenium 'light-switch' complexes can bind in the minor groove of DNA with five different binding modes.
Nucleic Acids Res., 44, 2016
|
|
6RII
 
 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Fourth structure of the series with 1200 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, FLUORIDE ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-24 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7RWG
 
 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | Descriptor: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
1F94
 
 | | THE 0.97 RESOLUTION STRUCTURE OF BUCANDIN, A NOVEL TOXIN ISOLATED FROM THE MALAYAN KRAIT | | Descriptor: | BUCANDIN | | Authors: | Kuhn, P, Deacon, A.M, Comoso, S, Rajaseger, G, Kini, R.M, Uson, I, Kolatkar, P.R. | | Deposit date: | 2000-07-06 | | Release date: | 2000-07-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The atomic resolution structure of bucandin, a novel toxin isolated from the Malayan krait, determined by direct methods.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5HQI
 
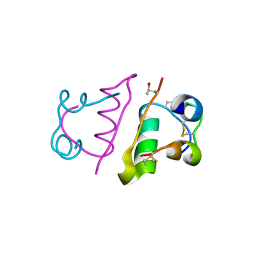 | | Insulin with proline analog HzP at position B28 in the T2 state | | Descriptor: | Insulin A-Chain, Insulin B-Chain | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
7WPW
 
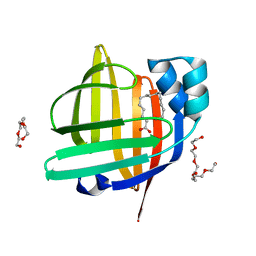 | | The 0.97 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with pentadecanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The 0.97 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with pentadecanoic acid
To Be Published
|
|
1XMK
 
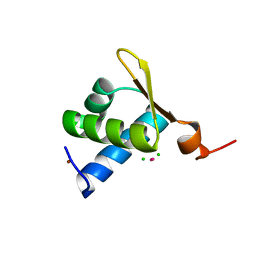 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
3U3H
 
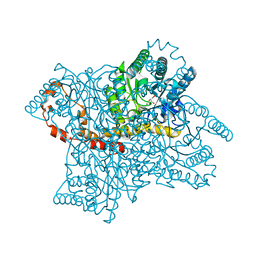 | | X-Ray Crystallographic Analysis of D-Xylose Isomerase-Catalyzed Isomerization of (R)-Glyceraldehyde | | Descriptor: | (2R)-propane-1,1,2,3-tetrol, (4R)-2-METHYLPENTANE-2,4-DIOL, FORMIC ACID, ... | | Authors: | Allen, K.N, Silvaggi, N.R, Toteva, M.M, Richard, J.P. | | Deposit date: | 2011-10-05 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Binding Energy and Catalysis by d-Xylose Isomerase: Kinetic, Product, and X-ray Crystallographic Analysis of Enzyme-Catalyzed Isomerization of (R)-Glyceraldehyde.
Biochemistry, 50, 2011
|
|
1ZUU
 
 | | Crystal structure of the yeast Bzz1 first SH3 domain at 0.97-A resolution | | Descriptor: | BZZ1 protein, MAGNESIUM ION, UNKNOWN ATOM OR ION | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structural genomics of yeast SH3 domains
To be Published
|
|
3LL2
 
 | |
485D
 
 | |
1G4I
 
 | | Crystal structure of the bovine pancreatic phospholipase A2 at 0.97A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Steiner, R.A, Rozeboom, H.J, de Vries, A, Kalk, K.H, Murshudov, G.N, Wilson, K.S, Dijkstra, B.W. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | X-ray structure of bovine pancreatic phospholipase A2 at atomic resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4MTU
 
 | | beta-Alanyl-CoA:Ammonia Lyase from Clostridium propionicum | | Descriptor: | Beta-alanyl-CoA:ammonia lyase 2, SULFATE ION, ZINC ION | | Authors: | Heine, A, Reuter, K. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High resolution crystal structure of Clostridium propionicum beta-alanyl-CoA:ammonia lyase, a new member of the "hot dog fold" protein superfamily.
Proteins, 82, 2014
|
|
6MU9
 
 | |
2XFR
 
 | | Crystal structure of barley beta-amylase at atomic resolution | | Descriptor: | 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
1BYI
 
 | | STRUCTURE OF APO-DETHIOBIOTIN SYNTHASE AT 0.97 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHASE | | Authors: | Sandalova, T, Schneider, G, Kaeck, H, Lindqvist, Y. | | Deposit date: | 1998-10-15 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of dethiobiotin synthetase at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3AKS
 
 | |
5DZE
 
 | | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with cellotetraose | | Descriptor: | beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, endo-glucanase | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2015-09-25 | | Release date: | 2016-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
5RCS
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 14 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
