4NQV
 
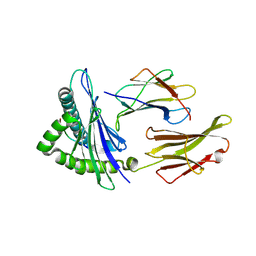 | |
6TXV
 
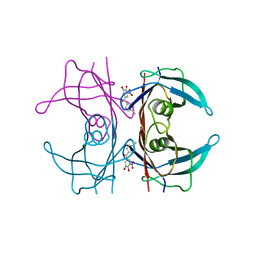 | | A25T Transthyretin structure in complex with Tolcalpone | | Descriptor: | Tolcapone, Transthyretin | | Authors: | Varejao, N, Reverter, D, Pinheiro, F, Pallares, I, Ventura, S. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tolcapone, a potent aggregation inhibitor for the treatment of familial leptomeningeal amyloidosis.
Febs J., 288, 2021
|
|
3Q4J
 
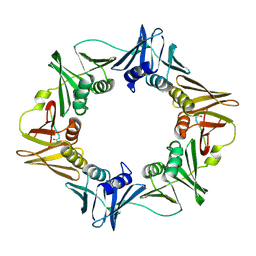 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
6RRA
 
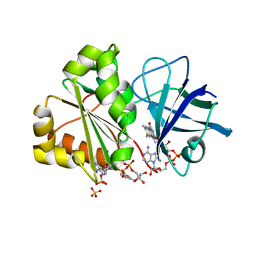 | | X-RAY STRUCTURE OF THE FERREDOXIN-NADP REDUCTASE FROM BRUCELLA OVIS IN COMPLEX WITH NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Martinez-Julvez, M, Taleb, V, Medina, M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Towards the competent conformation for catalysis in the ferredoxin-NADP+reductase from the Brucella ovis pathogen.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6RS2
 
 | | Structure of the Bateman module of human CNNM4. | | Descriptor: | Metal transporter CNNM4 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Gomez-Garcia, I, Oyenarte, I, Gimenez, P, Ereno-Orbea, J, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2019-05-21 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.694 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
2CYF
 
 | | The Crystal Structure of Canavalia Maritima Lectin (ConM) in Complex with Trehalose and Maltose | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Delatorre, P, Rocha, B.A.M, Sousa, E.P, Gadelha, C.A.A, Azevedo Jr, W.F, Cavada, B.S. | | Deposit date: | 2005-07-06 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a lectin from Canavalia maritima (ConM) in complex with trehalose and maltose reveals relevant mutation in ConA-like lectins
J.Struct.Biol., 154, 2006
|
|
3Q4K
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
2CXG
 
 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
2CY6
 
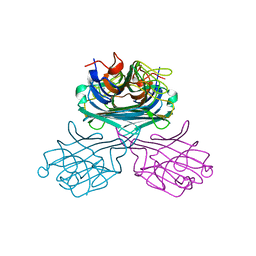 | | Crystal structure of ConM in complex with trehalose and maltose | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Cavada, B.S, Azevedo Jr, W.F, Delatorre, P, Rocha, B.A.M, Souza, E.P, Gadelha, C.A.A. | | Deposit date: | 2005-07-05 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lectin from Canavalia maritima (ConM) in complex with trehalose and maltose reveals relevant mutation in ConA-like lectins
J.Struct.Biol., 154, 2006
|
|
6OH8
 
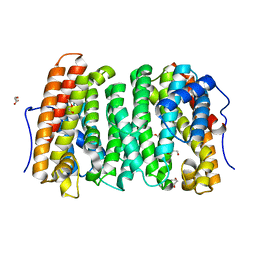 | |
6XQK
 
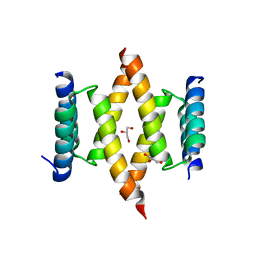 | | Crystal structure of the D/D domain of PKA from S. cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, cAMP-dependent protein kinase regulatory subunit | | Authors: | Larrieux, N, Gonzalez Bardeci, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of yeast regulatory subunit reveals key evolutionary insights into Protein Kinase A oligomerization.
J.Struct.Biol., 213, 2021
|
|
6OH6
 
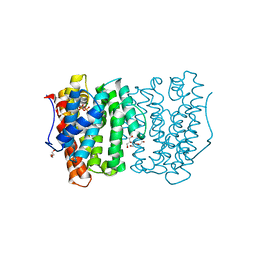 | |
2DVO
 
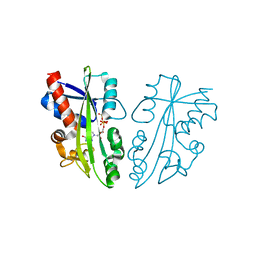 | |
1OJ7
 
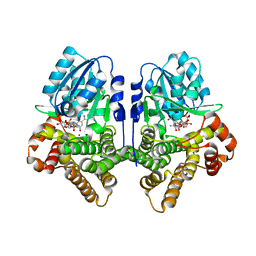 | | STRUCTURAL GENOMICS, UNKNOWN FUNCTION CRYSTAL STRUCTURE OF E. COLI K-12 YQHD | | Descriptor: | 5,6-DIHYDROXY-NADP, BORIC ACID, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Perrier, S, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-07-03 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E.Coli Alcohol Dehydrogenase Yqhd: Evidence of a Covalently Modified Nadp Coenzyme
J.Mol.Biol., 342, 2004
|
|
1OHG
 
 | | STRUCTURE OF THE DSDNA BACTERIOPHAGE HK97 MATURE EMPTY CAPSID | | Descriptor: | CHLORIDE ION, MAJOR CAPSID PROTEIN, SULFATE ION | | Authors: | Helgstrand, C, Wikoff, W.R, Duda, R.L, Hendrix, R.W, Johnson, J.E, Liljas, L. | | Deposit date: | 2003-05-26 | | Release date: | 2003-12-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Refined Structure of a Protein Catenane: The Hk97 Bacteriophage Capsid at 3.44A Resolution
J.Mol.Biol., 334, 2003
|
|
6XTF
 
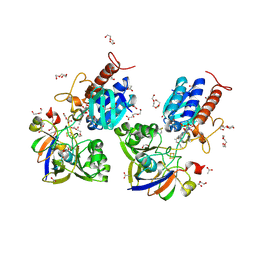 | | Crystal structure a Thioredoxin Reductase from Gloeobacter violaceus bound to its electron donor | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Buey, R.M, Gonzalez-Holgado, G, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria.
Plant Physiol., 186, 2021
|
|
1PEH
 
 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPNH1 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
6RR3
 
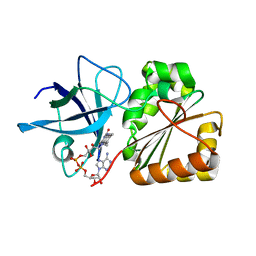 | |
3EK2
 
 | |
6V7A
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2657 | | Descriptor: | Hdac6 protein, N-hydroxy-4-[(1-methyl-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)methyl]benzamide, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.0874176 Å) | | Cite: | Spiroindoline-Capped Selective HDAC6 Inhibitors: Design, Synthesis, Structural Analysis, and Biological Evaluation.
Acs Med.Chem.Lett., 11, 2020
|
|
2C1E
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | Authors: | Grutter, M.G. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
3FGO
 
 | | Crystal Structure of the E2 magnesium fluoride complex of the (SR) Ca2+-ATPase with bound CPA and AMPPCP | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Laursen, M, Bublitz, M, Moncoq, K, Olesen, C, Moller, J.V, Young, H.S, Nissen, P, Morth, J.P. | | Deposit date: | 2008-12-08 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclopiazonic acid is complexed to a divalent metal ion when bound to the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 2009
|
|
3M4J
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
3CDS
 
 | |
3CZ1
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
